©Copyright 2018 GEOSCIENCE RESEARCH INSTITUTE
11060 Campus Street • Loma Linda, California 92350 • 909-558-4548

The AGEing Process: Rapid Post-Flood Intrabaraminic Diversification Caused by Altruistic Genetic Elements (Ages)
Todd Charles Wood
Assistant Professor, Center for Origins Research and Education
Bryan College, Dayton, Tennessee
P.O. Box 7604, Dayton TN 37321
wood@bryancore.org
WHAT THIS ARTICLE IS ABOUT
Transposition of genetic material is proposed as a solution to the problem of rapid post-Flood diversification of baramins. Mobile genetic elements (herein called Altruistic Genetic Elements, AGEs) fulfill three criteria for explaining post-Flood diversification: 1) they permanently alter the genome, 2) their alterations can be genespecific and beneficial, and 3) their beneficial activity was concentrated in the past. Thirteen biological phenomena are discussed in conjunction with evolution, creationist theories of diversification, and the AGEing hypothesis. All thirteen can potentially be explained under the AGEing mode, whereas only seven are explained by evolution and only one (intrabaraminic hybridization) is explained by previous creationist theories.
INTRODUCTION
Since 1941, modern creation biologists readily and openly admit the reality of speciation. In his book Fundamental Biology, Frank Marsh writes, “He who thinks that species (modern) of animals and plants remain fixed through successive generations, has but to examine nature’s record to discover his error. Variation is one of the most invariable laws in the biological world” (Marsh 1941, p 101). The Bible records clear discontinuities in living things, with fish, birds, plants, land animals, and man originating from separate acts of creation. Speciation must therefore be limited in scope and cannot be the source of the entire diversity of life. To reconcile these observations, Marsh coined the term baramin (“created kind”) to denote the boundary of variation. Within the baramin, speciation may occur, but no baramin ever evolves into another. Marsh spent most of his professional life promoting the baramin concept to his fellow creationists, but acceptance and application of his ideas has been slow.
Creationists who accepted Marsh’s concept were quick to adapt baramins to explain the capacity of the Ark (Whitcomb & Morris 1961, p 66ff; Woodmorappe 1996, p 5ff), with Jones (1973) proposing that no more than 2000 animals were on the Ark. More recently creationists have adopted and adapted Marsh’s terminology (Siegler 1974; Wise 1990; ReMine 1993, chap. 24; Robinson 1997), but actual organismal studies remain rare. The few baraminic analyses reveal large and highly variable baramins, lending support to Jones’s low estimate of the animal population on the Ark. Based on recent baraminology research, creationists have equated four mammalian families with baramins: Felidae (Mehlert 1995, Robinson & Cavanaugh 1998b), Camelidae (Wood et al. 1999), Equidae (Marsh 1947, p 177; Stein-Cadenbach 1993; Wood et al. 2001), and Canidae (Siegler 1974, Crompton 1993).
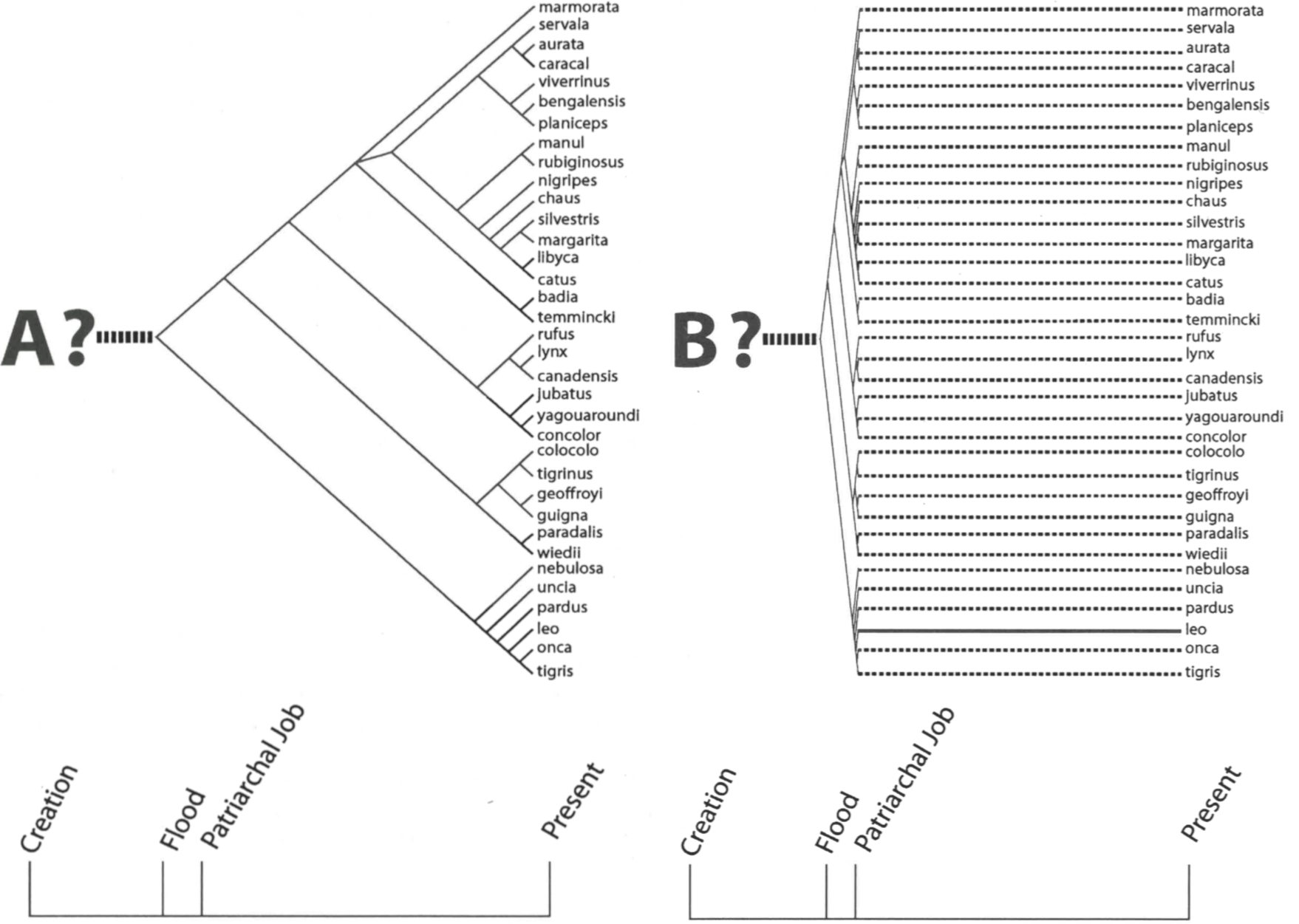
Species of these four mammalian baramins appear in early biblical passages (Table 1), highlighting the rapid intrabaraminic speciation that Wise (1994, 1995b) calls diversification. If we accept the creationist claim that only two of each (unclean) terrestrial baramin survived the Flood, then we must also accept a period of rapid intrabaraminic diversification, based on the early post-Flood appearance of modern species. For example, Robinson & Cavanaugh (1998b) follow Mehlert (1995) in assigning all extant cats to a single baramin. If they are correct, modern cat species have descended from a single pair of cats on the Ark. If it is a true representation of the history of the felid baramin, the cat phylogeny of Mattern & McLennan (2000) must therefore be a post-Flood history (Fig. 1A). Job mentions lions in four different passages (Table 1), and because Job is roughly a contemporary of Abraham, we may infer that lions first appeared within a few hundred years after the Flood. Since lions appear in a recent branch of Mattern & McLennan’s tree, we may infer that the diversification of cats should be compressed into a few hundred years after the Flood (Fig. 1B). A similar argument could be made for the other three mammalian baramins listed in Table 1.
The magnitude of variation required to produce all the cat (or dog or horse or camel) species in such a short time is assuredly unlike any variation or speciation we are presently observing. By inference from the studies cited above, we may make three observations about the cause(s) of intrabaraminic diversification. First, because of the apparent morphological stability of modern species, we infer that diversification must be caused by a permanent alteration of the organisms’ genomes. For example, tigers are always striped. They may vary in the saturation of the orange stripes, but every member of the species Panthera tigris possess black stripes. The stability of stripes in the tiger lineage argues for the stability of the genetic mechanism that produces them. Second, the rapidity of diversification would seem to eliminate neodarwinian mechanisms from the list of possible causes. To produce diversification so quickly, the mechanisms must specify the alterations in some way. Third, because we no longer observe speciation on the scale of intrabaraminic diversification, we infer that diversification has ceased. Consequently, proposed mechanisms of diversification ought to include a means of ending diversification after a limited amount of time.
Creationists have proposed a number of solutions to the problem of speciation, usually accepting some form of evolutionary mechanism. As John Morris notes, “Creationists agree with small, microevolutionary changes” (Morris 1999, p 6). While there is no reasonable doubt that Morris is correct, we must evaluate microevolutionary and speciation mechanisms in light of the three features just presented before invoking either as explanations of diversification. Creationist speciation mechanisms fall into three general categories, with several creationists accepting more than one category. The first category of speciation mechanism is actual change of the genetic material, as occurs with mutations or recombination. This mechanism is advocated by Jones (1982) and Marsh (1983). The second category of mechanism to explain speciation is hybridization, used by Marsh alone (Marsh 1983). By far the most popular category is the third: the fractionation of a heterozygous (genetically diverse) ancestral gene pool, or “heterozygous fractionation” for short. Siegler (1974), Morris (1974), Parker (1980), Scherer (1993), Batten (1996), and Wieland (1997) all use a type of heterozygous fractionation to explain speciation.
We may now compare these three categories of speciation mechanisms to the three features of intrabaraminic diversification to determine if the mechanisms are sufficient. First, genomic or genetic alteration is a permanent genetic change, so it meets one criterion for a diversification mechanism. Recombination and mutation occur too slowly today, so this mechanism alone fails to explain two out of three diversification features. Second, hybridization, in the sense that Marsh uses it, would be a permanent change, but it also still occurs today. Hybridization also requires an initial diversity of the baramin to produce lineages that can hybridize. Unclean mammal lineages immediately following the Flood would lack that diversity; thus, hybridization does not sufficiently explain diversification. Heterozygous fractionation has a modern example: dogs. It is accepted that all breeds of domestic dogs descended from the grey wolf, with most of the modern breeds arising within the last 500 years. Therefore, heterozygous fractionation appears to be capable of producing rapid change when coupled with extreme (artificial) selectional pressure. If we assume that residual post-Flood catastrophism provided strong selection for diversification, then the slow cessation of residual catastrophism would also yield a cessation of diversification.
Unfortunately, two aspects of heterozygous fractionation do not fit with diversification as outlined above. First, while breeding can generate phenotypic diversity in a short time, it fails to produce significant reproductive isolation to ensure the establishment of stable, persistent morphologies. As creationists are fond of pointing out, all dog varieties are still one species. Moreover, left to themselves, purebred dogs will quickly interbreed to produce mutts, which look increasingly wolf-like over multiple generations. Thus, the changes that result from heterozygous fractionation would probably not yield true species in the time available. An additional mechanism of reproductive isolation would be necessary. A second and more important difficulty of the heterozygous fractionation is the source of the initial heterozygosity. Some creationists unequivocally attribute initial heterozygosity to God’s direct creation (Siegler 1974, Weston & Wieland 1994). If we knew that baramins had gone through no bottlenecks in history, a directly created gene pool would explain the origin of heterozygosity. The Flood, however, produced a dramatic baraminic bottleneck for all land animal baramins, and probably for other baramins as well. After the Flood, the maximum number of alleles per locus for any land baramin would be four (unclean; or 14, clean). A survey of modern allelic diversity reveals a greater diversity than just four alleles per locus per baramin for many loci (e.g., see Tilley & Mahoney 1996). Again, another explanation must be invoked for the origin of these new alleles.
Could we combine explanations and thereby explain intrabaraminic diversification? For example, could mutations generate new alleles in the gene pool that then becomes fractionated? This approach might appear to solve the problems of the individual mechanisms, but in reality it merely compounds them. Invoking mutation to generate allelic diversity suffers from the same slow rate that mutation alone does. The mutation rate problem can only be alleviated by assuming a rapid rate of beneficial mutations that are not happening today, not by coupling mutations with heterozygous fractionation. From this brief survey, we conclude that speciation mechanisms as envisioned by creationists are inadequate to explain diversification. As early as 1974, Lammerts recognized this very problem. Instead of mechanisms that rely on natural selection, he proposed that God increased genetic variability for all baramins at Babel, coincident with the language confusion in humans (Lammerts & Howe 1974). He proposed this theory because he could think of no naturalistic explanation, although he remained open to them (Lammerts 1988).
Curiously, most creationists have all but ignored a speciation mechanism frequently used by botanists. Discovered by Barbara McClintock in the 1940s, mobile DNA can induce numerous phenotypic changes (McClintock 1950, Coen et al. 1989, Hartl 1989). Despite a prevailing negative view of mobile DNA as parasitic (Doolittle & Sapienza 1980, Orgel & Crick 1980), numerous researchers cite mobile DNA as an important evolutionary mechanism (see Flavell, Pearce & Kumar 1994; McDonald 1995; Lönnig & Saedler 1997; Kidwell & Lisch 1997, 2000; Wendel & Wessler 2000). Mobile DNA is a broad descriptor used for a number of DNA elements that have some ability to replicate and move in the genome independently from the normal DNA replication or recombination. Broadly speaking, mobile DNA may be divided into two classes. Class I elements transpose via an RNA intermediate and appear to be related to retroviruses. Class II elements transpose by a DNA intermediate using the enzyme transposase. Other DNA-based mobile DNA elements of unknown transposition mechanism have been suggested to form a third class. Most researchers would classify viruses as mobile DNA.
Mobile DNA, or transposable elements (TEs), display a number of features that could explain intrabaraminic diversification. For example, some TEs display a gene-specific distribution (Bureau & Wessler 1994, Mao et al. 2000), which could produce the specificity needed to rapidly alter genes. TE-induced mutations alter the genome in a mostly permanent fashion, although back mutations occasionally occur if the TE is excised. Most TEs are not currently active, but can be mobilized under special conditions, including hybridization (O’Neill, O’Neill & Graves 1998; Zhao et al. 1998) and climate change (Kalendar et al. 2000). The current inactivity of most TEs suggests that TE-induced phenotypic changes are also not occurring on a wide scale. All of these features are consistent with intrabaraminic diversification; however, some problems remain, namely the general lack of specificity of most TEs, which would result in a very slow rate of beneficial change. Because of this lack of specificity for most TEs, evolutionists tend to view TEs as sources of mutation upon which natural selection may act. Like any mutational mechanism mentioned above, it may suffer from a slow rate.
Although creationists have generally not used TEs to explain diversification, Brand & Gibson (1993) mentioned them in the context of a larger theory of speciation. In a discussion of potential origins of novel alleles for a broader heterozygous fractionation model, they briefly mention TEs then write “organisms were originally designed with an effective mechanism for increasing genetic variability, to meet changing conditions. These mechanisms may have suffered, after that time, from mutational damage, and are no longer as effective or as reliably beneficial as they originally were” (Brand & Gibson 1993). Thus, TEs might have lost their beneficial mutation capacity over time due to random mutations. The purpose of this article is to expand this unformed concept into a testable theory of intrabaraminic diversification. First, I will present the essentials of this new theory and how it differs from past creationist and evolutionist theories of speciation. Then I will outline thirteen biological evidences that could be explained by the new theory, only seven of which are explained by evolutionary speciation mechanisms and only one of which (intrabaraminic hybridization) is explained by current creationist theories.
TRANSPOSABLE ELEMENTS AND INTRABARAMINIC DIVERSIFICATION
Despite the unique ability of TEs to modify the genome rapidly, the low rate of beneficial mutations observed for most of these elements precludes their current utility in explaining speciation on the scale of diversification. Theoretical considerations regarding their initial created condition, however, reveal their value to diversification theory. If the original TEs had a higher rate of beneficial mutation, the speed at which TE-mediated diversification occurs would be greatly increased. The genomes of organisms must likewise be prepared in some way to receive the TE mutations. If the genome is not able to change appropriately, TE mutations can only be harmful. The cooperation of TEs and the genome to generate species diversity reflects a higher order complexity that seems inexplicable without divine design. God must design each organism’s genome to change in response to genomic alterations induced by TEs that were themselves designed to produce beneficial mutations. Without this initial design, TE-mediated diversification could not work, so to understand the true nature and purpose of TEs, we must view them in the context of a larger model of creation biology.
Although “transposable element” is a descriptive term, its connotation is far from neutral. The overwhelming majority of molecular biologists believe that TEs are genomic parasites that are ultimately harmful to their hosts (Orgel & Crick 1980, Doolittle & Sapienza 1980). My theory presented here differs substantially in that the effects of TEs were originally positive and historically have become either neutral or deleterious. Because of this important theoretical difference and because of the essentiality of design to my theory, I propose a new name for diversification-inducing transposable elements: Altruistic Genetic Elements (AGEs). This name will serve to distinguish the theoretical implications of my view and the genomic parasite theory. AGE-mediated diversification may simply be referred to as the AGEing process or AGEing for short.
How could AGEs modify the genome to generate species diversity in so little time? From modern examples of AGE-mediated genomic changes, we may infer one indirect and four direct methods of producing stable phenotypic changes in organismal lineages. The one indirect method involves the generation of novel recombination sites. AGEs and other repetitive DNA often serve as recombination sites within the genome (Himmelreich et al. 1997; Lohe et al. 2000; Cáceres, Puig & Ruiz 2001). While recombination does not directly alter the genetic content of the genome, it does promote allelic and genetic diversity in the population. Thus, AGE-induced recombination would be an indirect method of altering the phenotype of a lineage. The direct methods of altering the genome include gene disruption (Fig. 2B), AGE promoters (Fig. 2C), AGE enhancers (Fig. 2D), and actual gene transfer (Fig. 2E).
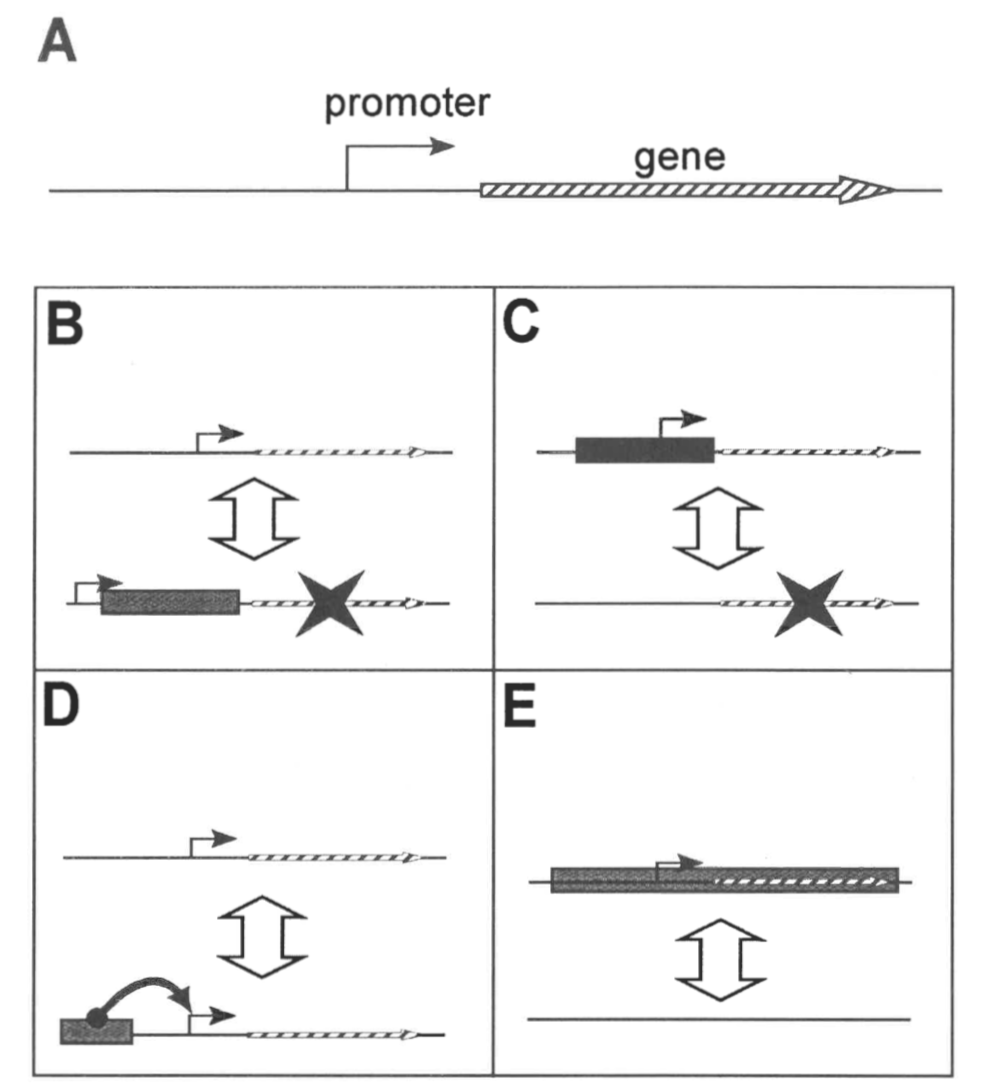
AGEs can inactivate genes if the AGE transposes into the coding or control region of the gene (Fig. 2B). In this manner, a gene may be shut off permanently at that locus. Conversely, if an organism was created with an AGE already present in a gene, that gene could potentially be activated upon excision of the AGE. Numerous mutations have been associated with AGE insertion (Collins & Gutman 1992; Arkhipova, Lyubomirskaya & Ilyin 1995; Chopra et al. 1999). As proposed above, the specifity of the AGE transposition would have to be very high to produce the rapid beneficial mutation rate needed for diversification. In the case of gene disruption by AGEs, if the transposition is specific for a certain sequence signature or a certain gene, each allele of a diploid or polyploid individual could be altered simultaneously upon activation of the appropriate AGE. Multiple origins of the same genetic alteration would promote the spread of the new alleles rapidly through the population, thereby avoiding potential problems associated with the slow rate of allele replacement in heterozygous fractionation models.
Some modern transposable elements are known or suspected to act as transcriptional promoters for genes (Fig. 2C) (Kim et al. 1989; White, Habera & Wessler 1994). If an AGE that carried a genetic promoter spliced in or out of the 5’ untranslated region of a gene, the expression pattern of that gene would be dramatically altered. As in the case of gene disruption, the mechanism could work in either of two ways. First, God could create a promoter-less gene and an AGE that was specific for inserting into that gene at a later date. In this way, the gene would become active. Second, God could create a gene with an AGE as its promoter that could excise itself from the gene at a later date. In this way, the gene would become inactive.
Although I know of only one example of a transposable element acting as a transcriptional enhancer (McDonald 1995), AGEs carrying enhancers could be a third mechanism to directly alter the phenotype of an organism (Fig. 2D). If God made AGEs that carried transcriptional enhancers, the movement of these AGEs could radically alter any number of gene expression patterns. Considering that enhancers often act at a physical distance from the gene (unlike the promoter), AGE enhancers could actually be a more potent way to change the phenotype of the host organismal lineage. As mentioned previously, the effects should be twofold: enhancing gene expression patterns upon insertion and reducing gene expression patterns upon excision.
Finally, AGEs may act by actually transposing complete genes between locations in a single genome or between organisms (Fig. 2E). True transposition would provide a complete copy of a new gene to an organism. Alternatively, transposition may work indirectly by moving a gene to a region of different recombination rate. From a baraminological perspective, it is relevant to ask whether AGEs may cross baraminic boundaries. Could an AGE created in one baramin insert and function in the genome of a different baramin? Theoretically, this would be possible if the AGE were designed to work in a different baramin. There are examples of AGEs transposing in the genomes of different baramins (Fischer, Wineholds & Plasterk 2001), and evidence from Drosophila and felids suggests that DNA can be transposed between individual species of the same baramin (Clark & Kidwell 1997; Jordan, Matyunina & McDonald 1999) and between individuals of different baramins (Robins & Samuelson 1992). Despite overwhelming evidence that horizontal transfer occurs, the mechanism by which DNA is transferred horizontally between species is not known.
These five potential mechanisms of genomic change induced by AGEs are given as examples only. Other mechanisms could be listed, including AGE-induced splicing variation (Hughes 2001) or gene promoter scrambling (Kloeckener-Gruissem & Freeling 1995). Whatever the mechanism, AGEing offers several potential inactivating mechanisms. First, in an unfallen world, AGEing would have ceased when the earth was filled and reproduction ceased. Second, in a fallen world, AGEing would have ceased as the beneficial AGE-induced mutation rate decreased as mutations in the AGEs themselves increased. Presumably, the features of the AGEs needed to transpose are less complex than the AGE features that induce beneficial mutation. If the beneficial mutation and transposition functions were strongly linked, deleterious mutations in one would necessarily affect the other, thus eliminating both transpositional and diversification activities. Without a strong link, mutations would quickly reduce or eliminate the beneficial mutation activity of AGEs without altering their transpositional success. Third, a higherlevel control mechanism, such as methylation, may have directly inactivated AGEs. In any case, AGEing would have ceased after a certain period of time.
One important implication of this theory is that AGEing probably took place during much of the pre-Flood period. Prior to the Flood, AGEing may have been sufficiently common to generate truly novel morphologies within as little as a single generation. The apparent species stasis with which we are so familiar would have been unusual in the pre-Flood world. AGE-mediated morphological variation also suggests that reproductive isolation may be a secondary consequence of diversification. Diversity of form precedes and probably contributes to isolation of reproductive lineages. As a result, other speciation mechanisms proposed by modern researchers may have little importance for the actual generation of biological diversity because of the heavy emphasis on reproductive isolation preceding the generation of new biological forms. To an evolutionist, reproductive isolation necessarily precedes diversity of form.
BIOLOGICAL FEATURES EXPLAINED BY AGEING
Although the AGEing process was originally conceived strictly to provide a genetic explanation of diversification, AGEs and the AGEing process have a high degree of explanatory power in many areas of biology. What follows is a brief discussion of thirteen biological phenomena that are potentially explained by the AGEing process. Further research will be necessary to demonstrate a link between the AGEing process and the phenomena discussed, but the potential of AGEing to explain each is very high. Of these evidences, twelve have never been explained within other creationist models. Evolutionists recognize only eleven of the features (two — diversification and high frequency of intrabaraminic hybridization — are uniquely creationist concerns). Of these eleven, only two are thought to be explained by a neodarwinian type of mutation/ selection, while five have random processes and historical contingency as their explanation, and four are unexplained (Table 2). Based on these evidences alone, the AGEing process is clearly the preferred model of intrabaraminic diversification; indeed, AGEing may well be the preferred model for speciation in general.
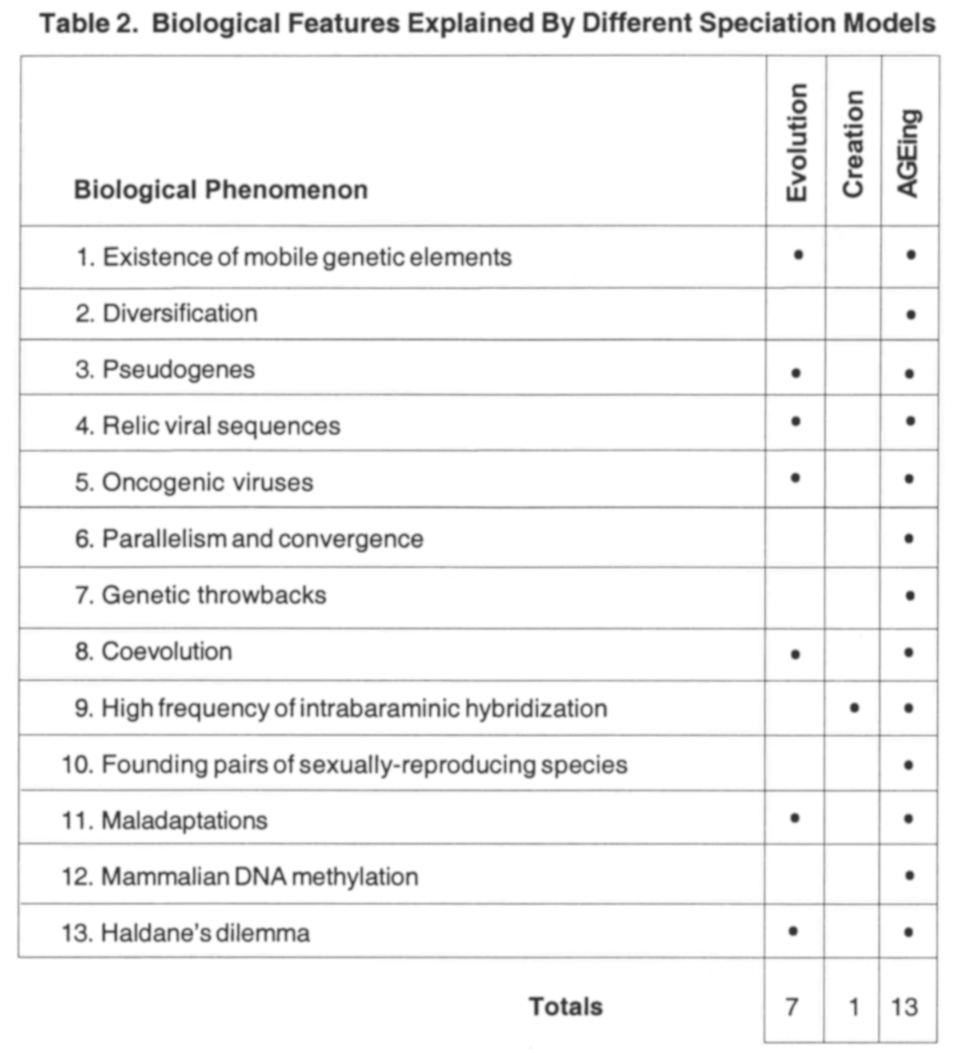
1. Existence of mobile genetic elements. As noted above, Doolittle & Sapienza (1980) and Orgel & Crick (1980) have provided the most popular evolutionary explanation for the persistence of mobile DNA by appealing to the selfishness of replicative DNA; however, they give no explanation for the origin of these sequences other than historical contingency, i.e., they just happened to evolve. Creationists have also wrestled with the origin of the mobile elements known as viruses, with some blaming viruses for the effects of the Fall, and others attempting to propose functions for viruses in an Edenic (Wise 1995a) and even postEdenic world (Bergman 1999). AGEing solves this conundrum neatly by not only providing a positive purpose for mobile DNA elements (and thus a purpose in God’s creation) but also explaining their ultimate failure and breakdown due to the effects of the Fall. The harmful effects of viruses today could be attributed to other mutations that changed the benignly reproducing AGEs into virulent pathogens, as Bergman has proposed (1999).
2. Rapid diversification. Although some creationists will undoubtedly try to draw parallels between intrabaraminic diversification and Eldredge & Gould’s punctuated equilibrium theory (1972), diversification is truly a separate phenomenon unto itself. No evolutionist would ever believe that all of horse, cat, or camel evolution occurred in less than five centuries, but this is the true essence of diversification. Diversification is speciation on a grand scale, at a rate evolutionists would scoff at. As noted above, some creationists have attempted to propose mechanisms to account for intrabaraminic speciation, but the AGEing process is the only model that explains the origin and cessation of diversification.
3. Pseudogenes. Pseudogenes come in two different forms: processed and unprocessed. In eukaryotes, processed pseudogenes lack introns and
Evolution Creation AGEing
typically have evidence of polyadenylation (addition of terminal adenines), as if they had been transcribed, processed to mRNA, reverse transcribed, and reinserted into the chromosome. Unprocessed pseudogenes are simply unexpressed normal genes. The failure of unprocessed pseudogenes to be expressed can result from mutations in the coding or control region of the gene. Evolutionists explain pseudogenes as abnormal retrotranspositions or as formerly active genes that have lost their ability to be expressed. In any case, evolutionists typically relegate pseudogenes to the class known as “junk DNA” and stress the randomness of the mutational processes that lead to their origins. In general, creationists have tried to explain pseudogenes by proposing functions just for pseudogenes without a broad consideration of intrabaraminic diversification (Gibson 1994).
The AGEing process provides one of two positive explanations for certain pseudogenes: 1) Some unprocessed pseudogenes may represent latent genes that were never activated or previously active genes that were inactivated by AGEs. 2) Processed pseudogenes may be the result of failed gene transfer by retrotransposon AGEs, after mutations began deteriorating their beneficial diversification function. These explanations may sound like the evolutionary explanations, but they differ in that AGE function is not an accident but an intentionally designed feature of AGEs.
4. Relic viral sequences. Genome sequencing projects have revealed a number of examples of genomic sequences of apparently viral origin, including phage sequences in the genome of the infectious bacterium Chlamydia trachomatis (Stephens et al. 1998) as well as hundreds of copies of retroviral sequences in mammalian genomes (Herniou et al. 1998). Evolutionists merely acknowledge their existence as the result of past viral infection, but have not yet proposed a positive explanation for their costly maintenance in the genomes. As with other types of “junk DNA,” creationists have only offered the hope that someday a function may be found for such sequences. In the AGEing model, relic viral sequences may play an unknown regulatory role; however, AGEing also allows the creationist to believe that these sequences may be currently non-functional relicts of the diversification process, possibly originating at the twilight of AGE activity.
5. Acutely and chronically oncogenic retroviruses. It is now common knowledge among cancer researchers that some viruses are among the most potent carcinogens. Acutely oncogenic (tumor-inducing) viruses can induce tumors in days to weeks, while chronically oncogenic viruses require longer latency periods of months to years (Peters 1989). By far the rarer of the two, acutely oncogenic retroviruses carry mutated versions of normal cellular genes called proto-oncogenes. Chronically oncogenic retroviruses alter the normal expression of endogenous genes through the enhancers and promoters present in the long terminal repeats (Peters 1989).
Evolutionists appeal to mutational accidents to explain the origin of acutely oncogenic retroviruses. Acutely oncogenic viruses are believed to acquire their proto-oncogenes during infection by a normal virus, mutation of that proto-oncogene to an oncogene due to the poor replicative fidelity of viruses, then reinfection of the host species (or even another species) and subsequent carcinogenesis (Alberts et al. 1994, p 1275). Chronically oncogenic viruses may be explained by a selfish DNA hypothesis. The cellular promoters and enhancers present in chronically oncogenic retroviruses arise from mutations favored by natural selection for viruses that most effectively utilize the cellular transcriptional machinery for their own reproduction.
Creationists have offered no explanation of oncogenic retroviruses. In the AGEing model, acutely oncogenic retroviruses can be explained as vestiges of transpositional AGEs, in which a formerly beneficial gene transfer function has become harmful due to mutation of the AGE. The mutagenesis of chronically oncogenic retroviruses may be explained in one of two ways: 1) mutations in the promoters or enhancers that inhibit their beneficial function, or 2) a breakdown of viral specificity, such that the virus either inserts where it should not or infects cells or species it should not. In each case, AGEing offers potential explanations of these unusual viruses.
6. Parallelism and convergence. Although most creationists might not realize it, parallelism and convergence does occur within and between baramins. The post-Flood development of the sabertooth characteristic in four different cat-like groups, the felids, nimravids, creodonts, and the marsupial thylacosmilids, is an example of such convergence (Simpson 1941, Radinsky & Emerson 1982). Within the evolution model, recent interest in the problem has helped to clarify the issues surrounding parallelism and convergence (Sanderson & Hufford 1996); nevertheless, both phenomena remain poorly explained. Anderson (1970) proposed that viruses may be responsible for the phenomenon, but his proposal has gone largely unnoticed.
Generally, creationists have used these phenomena to point to a common Designer of all living things, and rightly so, to some degree. Certainly, similarities between whales and land mammals must be relegated to the intentions of God the creator. The difficulty arises when true historical examples of parallelism or convergence occur within or between baramins (such as the sabertooth cats discussed above). In such cases, it is not clear what could cause such unusual development of traits.
The AGEing model provides a possible explanation by either true transposition or by the activation of similar latent genetic information in the same or different baramins. If the convergent trait was coded by a single gene, a transpositional AGE could insert that gene and therefore that trait in multiple organismal lineages, even simultaneously. Alternatively, we could relegate even historical convergence to God’s intentional design if we assume that the information required for the convergent traits was created in the genomes of different baramins in a latent state. Common AGEs could then activate the information later, producing convergence to the same traits. Even some of the most difficult examples of parallelism (e.g., ecosystem-wide adaptations) and convergence (e.g., mimicry) could be readily explained by the action of a common AGE or a group of similar AGEs.
7. Genetic throwbacks. Genetic throwbacks are organisms that express characteristics present only in ancestral species. An example is the two-toed horse documented by Othniel Marsh in the nineteenth century (Marsh 1879). Neither creationists nor evolutionists have provided an adequate explanation of genetic throwbacks. Evolutionary explanations fail because of the large timescales involved. It is reasonable to assume that over millions of years, the genetic information necessary to express ancestral traits should have degraded since it is unexpressed and therefore no longer subject to selection.
Creationists have difficulty with throwbacks for one of two reasons: either they believe in a very narrowly defined baramin (such as monodactyl and polydactyl horses in separate holobaramins — descended from separately created species), or they do not have a coherent model of how these traits have arisen in the first place. In the first case, genetic throwbacks become expression of traits of a separate baramin; in the second case, both the origin and reintroduction of the same trait are without explanation. In the AGEing model, genetic throwbacks are easily explained as the random reactivation of latent genetic information, either by an AGE that has not yet lost all diversification function or by some other mutational event. Because the timescales involved are much shorter than evolution, mutational degradation is not as important to explaining throwbacks.
8. Coevolution. Coevolution is generally described as the evolution of a species that depends on the evolution of another. Coevolutionary processes include predator-prey relationships, mutualistic symbioses, and parasite-host interactions. An excellent example of coevolution is the many species-specific adaptations of flowers and their pollinators. Evolutionists offer neodarwinian explanations of coevolution that are often quite complex, but creationists have never dealt with the issue, except to marvel at the “design” of such adaptations, even when speciesspecific adaptations are found within what is undoubtedly the same holobaramin (Clark 1965, Brauer 1972, Cornell 1975).
The AGEing process offers the potential explanation that AGEs transferred between the species coevolving could alter different specific genes within each species. Thus the adaptation of pollinator and flower is the manifestation of different responses of pre-designed genetic programs of the two species to the same AGE. The adaptation is “designed” in the sense of being planned by God, but is also the result of coevolution, in the sense that it developed after the originally created populations.
9. High frequency of intrabaraminic hybridization. Although the interspecific hybridization criterion for inclusion in a baramin has enjoyed a long history in creation biology (Marsh 1947, 1976; Scherer 1993), recent linguistic work by Williams has cast some doubt on the biblical basis of this criterion (Williams 1997). Nevertheless, based on alternative methods of identifying monobaramins (Robinson 1997; Robinson & Cavanaugh 1998a,b), the hybridization criterion does provide baraminologically useful information. AGEs provide a potential explanation as to why hybridization might be useful in baraminology. Since the changes induced by AGEs precede strict reproductive isolation, interspecific hybridization between members of the same baramin should still be possible. Thus, even species that appear to be very different, such as the llama and camel, are capable of hybridizing with much artificial help (Skidmore et al. 1999).
10. The founding pair of sexually-reproducing species. This critique is most often heard in conjunction with Goldschmidt’s macromutation theory (Sunderland 1988, p 115; Taylor 1991, p 164-165): if the random mutation that produces reproductive isolation necessary for speciation occurs in only one individual, how then are new species formed, since that individual is reproductively isolated? Evolutionists are forced to appeal to other types of reproductive isolation, such as geographic barriers followed by gradual mutation, to overcome this problem.
Unfortunately, this critique is also a problem for creationists who accept the very rapid speciation of diversification. If variation is sufficient to produce a wide range of morphological variation in a very short time, how can new reproductively-isolated species arise? The AGEing process provides a potential solution because diversity precedes reproductive isolation. Some AGE-related activities, such as transposition and high frequency of intrabaraminic hybridization, can also contribute to the origin of more than one member of a new “species.” AGEs that produce new morphologies can alter an entire population very quickly if they can be transferred horizontally among and between individual organisms. As noted above, the mechanism for such a change is unknown, but it remains a theoretical possibility. Alternatively, new morphologies can be passed through the baramin gene pool by hybridization between morphologically different parents. Hybridization would promote the general morphological uniformity of a population unless the population becomes geographically isolated during the “filling of the earth,” leading to other changes that induce reproductive isolation.
11. Maladaptations. Maladaptations are organismal characteristics that appear non-beneficial. A common example is the enormous antler spread of the extinct Irish elk Megaloceros giganteus (Gould 1974). Though proportionally consistent with the size of the Megaloceros body, an antler spread of up to 12 feet must have been cumbersome during feeding and running through the woods. Other than general appeals to allometry (changes in proportion during growth), or sexual selection (as in the case of Megaloceros), evolutionists provide few explanations for the persistence of maladaptations. Creationists wrestle with the very existence of maladaptations in a benevolently designed creation. The AGEing model could explain maladaptations as faulty AGEing activity due to the mutation of the AGE’s diversification function.
12. DNA methylation. In mammalian and plant genomes, the DNA sequence 5’-CG-3’ (CpG) is methylated on the cytosine base. Methylation (addition of a methyl group) has been implicated in the inactivation of the extra X chromosome of mammalian females (Heard, Clerc & Avner 1997), genomic imprinting (Bartolomei & Tilghman 1997), and in transcriptional suppression of mobile DNA (Yoder, Walsh & Bestor 1997). Since mobile DNA is viewed as a harmful genetic parasite by many evolutionists, some researchers propose that methylation evolved as a defense against the harmful effects of mobile DNA (Yoder, Walsh & Bestor 1997; Martienssen 1998). One objection to this hypothesis is that DNA methylation is a general mechanism for transcriptional inactivation used on genes other than TEs. Another objection is that the mammalian zygote is significantly demethylated. Alu retrotransposons inherited from the father and L1 retrotransposons inherited from the mother are both demethylated in the zygote, presumably allowing their transposition (Yoder, Walsh & Bestor 1997). Methylation reaches its normal adult levels about the time when cellular differentiation begins (Singal & Ginder 1999). Thus at the stage of ontogeny when the organism is most susceptible to mutational damage due to “selfish” mobile DNA, the organism drops much of its defense. A better solution to this problem is the AGEing process. In the AGEing model, methylation could serve a two-fold purpose: methylation ensures that AGE-induced changes do not occur during differentiation thus resulting in genetic mosaics (e.g., having a single antler or sabertooth), and demethylation in the zygote ensures that changes that occur at that stage are passed to all cells in the adult, including the germ line.
13. Haldane’s dilemma. The problem of the cost of substitution first brought to light by J.B.S. Haldane (1957) has been re-introduced to the creation-evolution debate by Walter ReMine (ReMine 1993, chap. 8). Briefly, Haldane’s dilemma states that the time required to substitute a new allele for an old one (the cost of substitution) is too long to allow for speciation to occur in the time required. ReMine uses the example of human evolution and shows that 7 million years is not enough time for the evolution of humans from the human/ape common ancestor. Evolutionists have never provided a solution to Haldane’s dilemma, instead focusing on model situations, such as the evolution of antibiotic resistance, where the (artificial) selection is unrealistically strong.
Since diversification also requires allele substitution, but on a much faster timescale, Haldane’s dilemma applies even more strictly to creationist models of speciation, particularly heterozygous fractionation. Besides the strong natural selection proposed by the evolutionists, another solution to Haldane’s dilemma is multiple introductions of the same new allele. In a random neodarwinian speciation process, multiple evolution of the same novel allele is extremely unlikely, but in the AGEing process, it is predicted. Alternatively, in the case of transpositional AGEs, new alleles can spread through an entire population like an infection. In either case, Haldane’s dilemma can be overcome by the action of AGEs.
PREDICTIONS
The hallmark of all good scientific models is testable predictions. Past discussions of creation biology have focused primarily on proposed features of divine design, without the inclusion of any testable predictions. The AGEing process clearly distinguishes itself from much of previous creation biology by making numerous testable predictions. Three general predictions are discussed below, with preliminary confirmation of each. More research will be necessary to demonstrate successful and convincing predictions of the AGEing model.
Prediction #1: The difference between two cobaraminic species will be found in AGEs. If AGEs are the causative agents of speciation, it logically follows that genomic differences between species of the same baramin will be largely restricted to AGEs and AGE-induced genetic alterations. Positive evidence for this prediction has been observed in numerous modern genome projects. Comparison of preliminary results of the genome sequence of the nematode Caenorhabditis briggsae to the completed genome of Caenorhabditis elegans revealed that the gene content and order is well-conserved, but the intergenic and intron sequences are shorter in C. briggsae. The difference in length of these regions is due to the absense of certain repetitive and transposable elements in C. briggsae that are present in C. elegans (Blaxter 1998).
Further confirmation of this prediction came from the sequence of the mycoplasma urinary tract pathogen Ureaplasma urealyticum (Glass et al. 2000). Based on the well-conserved gene order between Mycoplasma genitalium and Mycoplasma pneumoniae, I proposed a model for the origin of pathogenesis in the mycoplasmas that invoked genomic reduction due primarily to faulty recombination (Wood 2001). The genome sequence of U. urealyticum revealed a far greater rearrangement of genes than I expected to see based on observations of the two sequenced Mycoplasma genomes. Despite being the most closely related species to M. genitalium and M. pneumoniae, the gene order in U. urealyticum and the two Mycoplasma species is very poorly conserved. The genomic reduction of the mycoplasmas is more complicated than simple faulty recombination can account for; however, the presence of six transposons in the U. urealyticum genome that are absent in both Mycoplasma genomes could account for the genomic rearrangement between these very closely related bacterial species (Glass et al. 2000). Since transposons are AGEs, the gene order in the Ureaplasma and Mycoplasma genomes may confirm the first prediction of the AGEing process.
Finally, a number of Miniature Inverted-repeat Transposable Elements (MITEs) are known from studies of the genes of rice (Oryza sativa). A recent survey of MITEs in 73,362 genome survey sequences of O. sativa revealed that MITEs show a species-specific frequency (Mao et al. 2000). MITEs identified in African species of rice (O. longistaminata and O. glaberrima) occur with much less frequency in the genome survey sequences than MITEs originally identified in the genome of O. sativa.
Prediction #2: In the actual chromosome sequence, some AGEs should be physically associated with genes responsible for species specific traits. Although AGE enhancers can act at a distance from the gene they influence, in general, it is reasonable to expect AGEs to be associated closely with the genes they affect. This means that in a chromosomal sequence, we should find repetitive DNA, pseudogenes, and mobile elements positioned in proximity to genes expressing speciesspecific traits. The converse of this prediction is that AGEs will not be associated strongly with genes required for cellular survival, such as genes used in metabolism, protein synthesis, DNA replication, or RNA transcription.
This prediction finds a preliminary confirmation in the recently completed genome sequence of C. elegans. One of the first studies done on the genome was a comparison of the genes of C. elegans to the genes of the yeast Saccharomyces cerevisiae. Genes shared by both of these organisms are most likely to be those genes required for cellular processes; genes that are unique to each organism are the genes that give them their unique organismal characteristics. It was soon discovered that the genes common to C. elegans and S. cerevisiae tended to cluster toward the center of each chromosome, whereas the repetitive DNA found on the C. elegans autosomes were found on the arms of the chromosomes, away from the center (C. elegans Sequencing Consortium 1998). Thus the cellular process genes are not positionally associated with the AGEs, as the AGEing model predicts. Clearly, more studies will be necessary to systematize the AGE/gene association and to categorize the genes that may be affected by adjacent AGEs.
Prediction #3: Populations living immediately after the Flood were more adaptable than populations living now. Since AGEs have become less active over time due to mutation, the biological consequence is that modern baramins are less adaptable to adverse environmental changes. With AGEs active, new traits were routinely introduced into each population of organisms living just after the Flood, strongly influencing their ability to move from environment to environment. This adaptability no doubt aided in their survival in the tumultuous residual catastrophism of the post-Flood world. Additionally, this loss of adaptability may well explain the mysteriously high extinction rates of modern species.
Since there have been a number of drastic environmental changes recorded in the post-Flood fossil record (including the end of the Ice Age), the adaptability of baramins could be measured as the number of species per baramin before and after the environment change. The AGEing model predicts that fewer species per baramin will survive environmental changes as time after the Flood progresses.
SUMMARY
I have presented a model of intrabaraminic diversification that is conceptually simple and fulfills all the requirements for a theory of diversification. Rapid intrabaraminic diversification is attributed to Altruistic Genetic Elements (AGEs), which are designed by God to cause permanent, beneficial genomic changes. I presented thirteen general biological evidences that can be explained by the AGEing process as well as three specific predictions of the AGEing process that can be tested. The AGEing process also has applicability to diverse problems within creation biology. Space precludes detailed discussion of the application of the theory, but problems that could be addressed by AGEing include marsupial biogeography, dinosaur extinction, extreme human longevity, and survival of freshwater fish through the Flood.
With a theoretical framework such as the AGEing model of diversification, creation biology may finally blossom as a full-fledged discipline within the creation model. No longer do creation biologists need to restrict their work to theorizing over individual puzzles or marveling at ill-defined “design.” With a proposed model, creation biology can turn to the task of testing and refinement. While it is certainly possible that the AGEing process may ultimately be rejected, the model itself provides a plan of research that is indispensable to the growth of creation biology as a quality science.
ACKNOWLEDGMENTS
A preliminary version of this paper was presented at Baraminology 99, Liberty University, August 5-7, 1999. I would like to thank Kurt Wise for his careful reading and critique of an earlier draft of this paper, John Mark Reynolds, Stephanie Hartz, and an unnamed reviewer for very helpful discussions and suggestions.
LITERATURE CITED
Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. 1994. Molecular biology of the cell. 3rd ed. NY: Garland Publishing.
Anderson NG. 1970. Evolutionary significance of virus infection. Nature 227:13461347.
Arkhipova IR, Lyubomirskaya NV, Ilyin YV. 1995. Drosophila retrotransposons. Austin, TX: R.G. Landes Co.
Bartolomei MS, Tilghman SM. 1997. Genomic imprinting in mammals. Annual Review of Genetics 31:493-525.
Batten D. 1996. Dogs breeding dogs? That’s not evolution! Creation Ex Nihilo 18(2):21-23.
Bergman J. 1999. Did God make pathogenic viruses? Creation Ex Nihilo Technical Journal 13:115-125.
Blaxter M. 1998. Caenorhabditis elegans is a nematode. Science 282:2041-2046. Brand LR, Gibson LJ. 1993. An interventionist theory of natural selection and biological change within limits. Origins 20:60-82.
Brauer OL. 1972. The Smyrna fig requires God for its production. Creation Research Society Quarterly 9:129-131.
Bureau TE, Wessler SR. 1994. Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proceedings of the National Academy of Sciences (USA) 91:1411-1415.
Cáceres M, Puig M, Ruiz A. 2001. Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Research 11:1353-1364.
The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012-2018.
Chopra S, Brendel V, Zhang J, Axtell JD, Peterson T. 1999. Molecular characterization of a mutable pigmentation phenotype and isolation of the first active transposable element from Sorghum bicolor. Proceedings of the National Academy of Sciences (USA) 96:15330-15335.
Clark HW. 1965. The plants will teach you. Creation Research Society Quarterly 2:3-5.
Clark JB, Kidwell MG. 1997. A phylogenetic perspective on P transposable element evolution in Drosophila. Proceedings of the National Academy of Sciences (USA) 94:11428-11433.
Coen ES, Robbins TP, Almeida J, Hudson A, Carpenter R. 1989. Consequences and mechanisms of transposition in Antirrhinum majus. In Berg DE, Howe MM, editors. Mobile DNA. Washington DC: American Society for Microbiology, p 413-436.
Collins CM, Gutman DM. 1992. Insertional inactivation of an Escherichia coli urease gene by IS3411. Journal of Bacteriology 174:883-888.
Cornell AB. 1975. The moccasin flower was designed. Creation Research Society Quarterly 12:139-140.
Crompton NEA. 1993. A review of selected features of the family Canidae with reference to its fundamental taxonomic status. In Scherer S, editor. Typen des Lebens. Berlin: Pascal-Verlag, p 217-224.
Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm, and genome evolution. Nature 284:601-603.
Eldredge N, Gould SJ. 1972. Punctuated equilibria: an alternative to phyletic gradualism. In Schopf TJM, editor. Models in Paleobiology. San Francisco: Freeman, Cooper and Co., p 82-115.
Fischer SEJ, Wineholds E, Plasterk RHA. 2001. Regulated transposition of a fish transposon in the mouse germ line. Proceedings of the National Academy of Sciences (USA) 98:6759-6764.
Flavell AJ, Pearce SR, Kumar A. 1994. Plant transposable elements and the genome. Current Opinion in Genetics and Development 4:838-844.
Gibson LJ. 1994. Pseudogenes and origins. Origins 21:91-108.
Glass JI, Lefkowitz EJ, Glass JS, Heiner CR, Chen EY, Cassell GH. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762.
Gould SJ. 1974. The origin and function of “bizarre” structures: antler size and skull size in the “Irish elk,” Megaloceros giganteus. Evolution 28:191-220.
Haldane JBS. 1957. The cost of natural selection. Journal of Genetics 55:511-524.
Hartl DL. 1989. Transposable element mariner in Drosophila species. In Berg DE, Howe MM, editors. Mobile DNA. Washington DC: American Society for Microbiology, p 531-536.
Heard E, Clerc P, Avner P. 1997. X-chromosome inactivation in mammals. Annual Review of Genetics 31:571-610.
Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. 1998. Retroviral diversity and distribution in vertebrates. Journal of Virology 72:5955-5966.
Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Research 25:701-712.
Hughes DC. 2001. Alternative splicing of the human VEGFGR-3/FLT4 gene as a consequence of an integrated human endogenous retrovirus. Journal of Molecular Evolution 53:77-79.
Jones AJ. 1973. How many animals in the Ark? Creation Research Society Quarterly 10:102-108.
Jones AJ. 1982. The genetic integrity of the “kinds” (baramins): a working hypothesis. Creation Research Society Quarterly 19:13-18.
Jordan IK, Matyunina LV, McDonald JF. 1999. Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proceedings of the National Academy of Sciences (USA) 96:12621-12625.
Kalendar R, Transkanen J, Immonen S, Nevo E, Schulman AH. 2000. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences (USA) 97:6603-6607.
Kidwell MG, Lisch D. 1997. Transposable elements as sources of variation in animals and plants. Proceedings of the National Academy of Sciences (USA) 94:7704-7711.
Kidwell MG, Lisch D. 2000. Transposable elements and host genome evolution. Trends in Ecology and Evolution 15:95-99.
Kim JH, Yu CY, Bailey A, Hardison R, Shen CK. 1989. Unique sequence organization and erythroid cell-specific nuclear factor-binding of mammalian theta 1 globin promoters. Nucleic Acids Research 17:5687-5700.
Kloeckener-Gruissem, B, Freeling M. 1995. Transposon-induced promoter scrambling: a mechanism for the evolution of new alleles. Proceedings of the National Academy of Sciences (USA) 92:1836-1840.
Lammerts WE. 1988. Concerning disjunct populations of mammals and plants. Creation Research Society Quarterly 25:126-128.
Lammerts WE, Howe GF. 1974. Plant succession studies in relation to micro-evolution. Creation Research Society Quarterly 10:208-228.
Lohe AR, Timmons C, Beerman I, Lozovskaya ER, Hartl DL. 2000. Self-inflicted wounds, template-directed gap repair, and a recombination hotspot: effects of the mariner transposase. Genetics 154:647-656.
Lönnig W-E, Saedler H. 1997. Plant transposons: contributors to evolution? Gene 205:245-253.
Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Woo S-S, Sasinowski M, Presting G, Frisch D, Goff S, Dean RA, Wing RA. 2000. Rice transposable elements: a survey of 73,000 sequence-tagged connectors. Genome Research 10:982-990.
Marsh FL. 1941. Fundamental biology. Lincoln, NE: Published by author.
Marsh FL. 1947. Evolution, creation, and science. 2nd ed. Washington DC: Review and Herald Publishing Association.
Marsh FL. 1976. Variation and fixity in nature. Mountain View, CA: Pacific Press Publishing Association.
Marsh FL. 1983. Genetic variation, limitless or limited? Creation Research Society Quarterly 19:204-206.
Marsh OC. 1879. Polydactyle horses, recent and extinct. American Journal of Science 17:499-505.
Martienssen R. 1998. Transposons, DNA methylation and gene control. Trends in Genetics 14:263-264.
Mattern MY, McLennan DA. 2000. Phylogeny and speciation of felids. Cladistics 16:232253.
McClintock B. 1950. The origin and behavior of mutable loci in maize. Proceedings of the National Academy of Sciences (USA) 36:344-355.
McDonald JF. 1995. Transposable elements: possible catalysts of organismic evolution. Trends in Ecology and Evolution 10:123-126.
Mehlert AW. 1995. On the origin of cats and carnivory. Creation Ex Nihilo Technical Journal 9:106-120.
Morris HM. 1974. Diversity of opinions found in creationism. Creation Research Society Quarterly 11:173-174.
Morris JD. 1999. Dinosaurs, the lost world, & you. Green Forest, AR: Master Books.
O’Neill RJW, O’Neill MJ, Graves JAM. 1998. Undermethylation associated with retroelement activation and chromosome remodeling in an interspecific mammalian hybrid. Nature 393:68-72.
Orgel LE, Crick FHC. 1980. Selfish DNA: the ultimate parasite. Nature 284:604-607.
Parker GE. 1980. Creation, mutation, and variation. ICR Impact 89.
Peters G. 1989. Oncogenes at viral integration sites. In Glover DM, Hames BD. Oncogenes. Oxford: Oxford Unversity Press, p 23-66.
Radinsky L, Emerson S. 1982. The late, great sabertooths. Natural History 91(4):50-57.
ReMine WJ. 1993. The biotic message: evolution vs. message theory. St. Paul, MN: St. Paul Science.
Robins DM, Samuelson LC. 1992. Retrotransposons and the evolution of mammalian gene expression. Genetica 86:191-201.
Robinson DA. 1997. A mitochondrial DNA analysis of the testudine apobaramin. Creation Research Society Quarterly 33:262-272.
Robinson DA, Cavanaugh DP. 1998a. A quantitative approach to baraminology with examples from the Catarrhine primates. Creation Research Society Quarterly 34:196208.
Robinson DA, Cavanaugh DP. 1998b. Evidence for a holobaraminic origin of the cats. Creation Research Society Quarterly 35:2-14.
Sanderson MJ, Hufford L, editors. 1996. Homoplasy: the recurrence of similarity in evolution. NY: Academic Press.
Scherer S. 1993. Basic types of life. In Scherer S, editor. Typen des Lebens. Berlin: Pascal Verlag, p 11-30.
Siegler HR. 1974. The magnificence of kinds as demonstrated by Canids. Creation Research Society Quarterly 11:94-97.
Simpson GG. 1941. The function of saber-like canines in carnivorous mammals. American Museum Novitates 1130.
Singal R, Ginder GD. 1999. DNA methylation. Blood 93:4059-4070.
Skidmore JA, Billah M, Binns M, Short RV, Allen WR. 1999. Hybridizing old and new world camelids: Camelus dromedaries x Lama guanicoe. Proceedings of the Royal Society of London B266:649-656.
Stein-Cadenbach H. 1993. Hybriden, chromosomen und artbildung bei pferden (Equidae). In Scherer S, editor. Typen des Lebens. Berlin: Pascal-Verlag, p 225-244.
Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchel W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759.
Sunderland LD. 1988. Darwin’s enigma. Freen Forest, AR: Master Books.
Taylor IT. 1991. In the minds of men. 3rd ed. Minneapolis, MN: TFE Publishing.
Tilley SG, Mahoney MJ. 1996. Patterns of genetic differentiation in salamanders of the Desmognathus ochrophaeus complex (Amphibia: Plethodontidae). Herpetological Monographs 10:1-42.
Wendel JF, Wessler SR. 2000. Retrotransposon-mediated genome evolution on a local ecological scale. Proceedings of the National Academy of Sciences (USA) 97:62506252.
Weston P, Wieland C. 1998. Bears across the world.... Creation Ex Nihilo 20(4):29-31.
Whitcomb JC, Morris HM. 1961. The Genesis flood. Phillipsburg, NJ: Presbyterian and Reformed Publishing.
White SE, Habera LF, Wessler SR. 1994. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proceedings of the National Academy of Sciences (USA) 91:1179211796.
Wieland C. 1994. Speciation conference brings good news for creationists. Creation Ex Nihilo Technical Journal 11:135-136.
Williams PJ. 1997. What does min mean? Creation Ex Nihilo Technical Journal 11:344352.
Wise KP. 1990. Baraminology: a young-earth creation biosystematic method. In Walsh RE, Brooks CL, editors. Proceedings of the Second International Conference on Creationism, Vol. 2. Pittsburgh: Creation Science Fellowship, p 345-360.
Wise KP. 1994. Australopithecus ramidus and the fossil record. Creation Ex Nihilo Technical Journal 8:160-165.
Wise KP. 1995a. It matters where you start. In Land RD, Moore LA, editors. Life at Risk: The Crisis in Medical Ethics. Nashville, TN: Broadman & Holman.
Wise KP. 1995b. A note on new australopithecines. Creation Ex Nihilo Technical Journal 9:167.
Wood TC. 2001. Genome decay in the mycoplasmas. ICR Impact 340.
Wood TC, Williams PJ, Wise KP, Robinson DA. 1999. Summaries on Camelid Baraminology. In Baraminology 99. Baraminology Study Group, p 9-18. Available online at http://www.bryancore.org/bsg/
Wood TC, Wise KP, Cavanaugh DP. 2001. Pattern recognition analysis of fossil horses confirms the reality of the stratomorphic series. In Helder M, editor. Discontinuity: Understanding Biology in the Light of Creation. Baraminology Study Group, p 34. Available online at http://www.bryancore.org/bsg/
Woodmorappe J. 1996. Noah’s Ark: a feasibility study. Santee, CA: Institute for Creation Research.
Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends in Genetics 13:335-340.
Zhao X, Si Y, Hanson RE, Crane CF, Price HJ, Stelly DM, Wendel JF, Paterson AH. 1998. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Research 8:479-492.