©Copyright 2018 GEOSCIENCE RESEARCH INSTITUTE
11060 Campus Street • Loma Linda, California 92350 • 909-558-4548

A BARAMINOLOGICAL ANALYSIS OF SUBTRIBE FLAVERIINAE (ASTERACEAE: HELENIEAE) AND THE ORIGIN OF BIOLOGICAL COMPLEXITY
Todd Charles Wood
wood@bryancore.org
Assistant Professor, Center for Origins Research and Education
Bryan College, P.O. Box 7731
Dayton, TN, 37321;
and
David P. Cavanaugh
27329 Alberta Drive,
Harvest, AL 35749
WHAT THIS ARTICLE IS ABOUT
The subtribe Flaveriinae (Asteraceae: Helenieae) includes a number of plant species sometimes called yellowtops, glowworts, and false broomweed. Different species of this subtribe differ in their chemical pathways involved in photosynthesis. Some species use a system known as C3 photosynthesis, some use a system known as C4 photosynthesis, and others display characteristics intermediate between the two. The authors apply a creationist research method known as baraminology to determine whether the species might have been created separately, or whether they may have descended from a single created ancestral species. They present a large amount of evidence that suggests the entire subtribe belongs to a single lineage which includes additional species not included in the study. The evidence also implies that the originally created ancestor used C3 photosynthesis, and that the C4 photosynthesis present in some species emerged since the creation. The characteristics of the intermediate species and the genetics of C4 species support the hypothesis that latent genetic information may have been present in the ancestor, and activated during post-Flood diversification of the group, possibly through a mechanism called Altruistic Genetic Elements.
INTRODUCTION
Creationists have long speculated whether the "created kind" can be approximated by a traditional Linnean classification level (Marsh 1976), with several creationists proposing that kinds may be equivalent to families (Jones 1972, Siegler 1978, Woodmorappe 1996). In general, these speculations are based largely on hybridization studies of vertebrates or on the lists of organisms in Leviticus. Although the family/kind approximation may be adequate for mammals and birds, for many other types of organisms this estimation may certainly be incorrect. Flowering plant families frequently contain thousands of species. For example, the Iridaceae contains 1500 species, the Melastomataceae 4000 species, and the Euphorbiacea 7500 species (Cronquist 1981). The largest plant family is Asteraceae, with estimates ranging from 20,000 species in 1100 genera (Cronquist 1981) to 30,000 species in 2500 genera (Kim and Jansen 1995). While it is certainly possible that the Asteraceae could be a single created kind, its species diversity would be without parallel among the vertebrates. Thus, the magnitude of variation in the plants should be cause for skepticism in too quickly equating "family" with "created kind."
As a preliminary case study of created kinds in plants, we present here a review of the taxonomy and phylogeny of the plants of subtribe Flaveriinae (Asteraceae). Wise's (1992) method of baraminology evaluates additive and subtractive evidence separately in an attempt to approximate the complete membership of the created kind, or holobaramin. Additive evidence suggests that two species are genealogically related and is used to delineate monobaramins. Properly understood, monobaramins are simply a group of species which share a common ancestor; the group may be monophyletic or paraphyletic. By revealing a common developmental mode, the traditional hybridization criterion (Marsh 1976) is a source of additive evidence. Subtractive evidence suggests that two species or groups have a separate ancestry and is used to define apobaramins. An apobaramin is any group of species that are separated from all other species by a phylogenetic discontinuity; the apobaramin may contain one or more holobaramins. Novel biological features, such as flight in bats, are frequently used as subtractive evidences. Additive and subtractive evidence can also be evaluated together using the ANalysis Of PAttern (ANOPA) method, which is effective in identifying significant continuity and discontinuity in phylogenetic data (Cavanaugh and Sternberg, in prep).
Currently, two different definitions of subtribe Flaveriinae exist. Flaveriinae sensu stricto includes only three genera, Sartwellia (4 spp., Turner 1971), Haploësthes (3 spp., Turner 1975), and Flaveria (21 spp., Powell 1978); while Flaveriinae sensu lato includes those three genera and the genera Clappia, Jaumea, Pseudoclappia, and Varilla (Karis and Ryding 1994). For simplicity's sake, herein we will use the strict definition of Flaveriinae with only three genera, referring to Flaveriinae sensu lato where necessary.
The plants of each genus of Flaveriinae prefer gypsiferous soils in arid climates. Sartwellia and Haploësthes are both found in overlapping ranges in New Mexico, Texas, and Mexico (Turner 1971, 1975). The range of Flaveria extends from the southwestern United States through Central America into South America and includes Florida and several Caribbean islands. Isolated Flaveria species are also found in Africa, India, and Australia (Bremer 1994, Powell 1978). Like all Asteraceae, the Flaveriinae reproduce by miniature flowers, which are aggregated into heads resembling larger single flowers, and one-seeded achenes (such as the familiar, striped shells containing the edible sunflower seeds). Unlike other Asteraceae, Flaveriinae contain large amounts of sulfated flavonoids, presumably as an adaptation to or consequence of the saline, sulfate-rich niches they occupy (Powell 1978).
The genus Flaveria is surprisingly diverse, containing annual and perennial species as well as woody and herbaceous species. Most remarkable of all, Flaveria is one of only a handful of genera that contain species that photosynthesize by the C3 and C4 pathways as well as various C3-C4 intermediates.
To review, the C3 plants fix carbon dioxide into a three-carbon compound, 3-phosphoglycerate (3-PGA). This reaction is catalyzed by the enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisco). 3-PGA then proceeds through a series of reactions known as the Calvin cycle, whereby CO2 from the atmosphere is incorporated into sugars. True C4 plants are characterized by the presence of Kranz anatomy, a ring of specialized bundle sheath cells (BSC) that surround the veins of the leaf. CO2 is fixed in the regular mesophyll cells (MC) of the leaf into a four-carbon compound, oxaloacetate or malate, catalyzed by phosphoenolpyruvate carboxylase (PEPC). These four-carbon compounds ultimately are transported to the BSC where they are broken down, releasing carbon dioxide, which is then fixed into 3-PGA by the enzyme rubisco (Figure 1). The advantages of C4 photosynthesis include better nitrogen-use efficiency and lower photorespiration than in C3 plants (Edwards and Walker 1983).
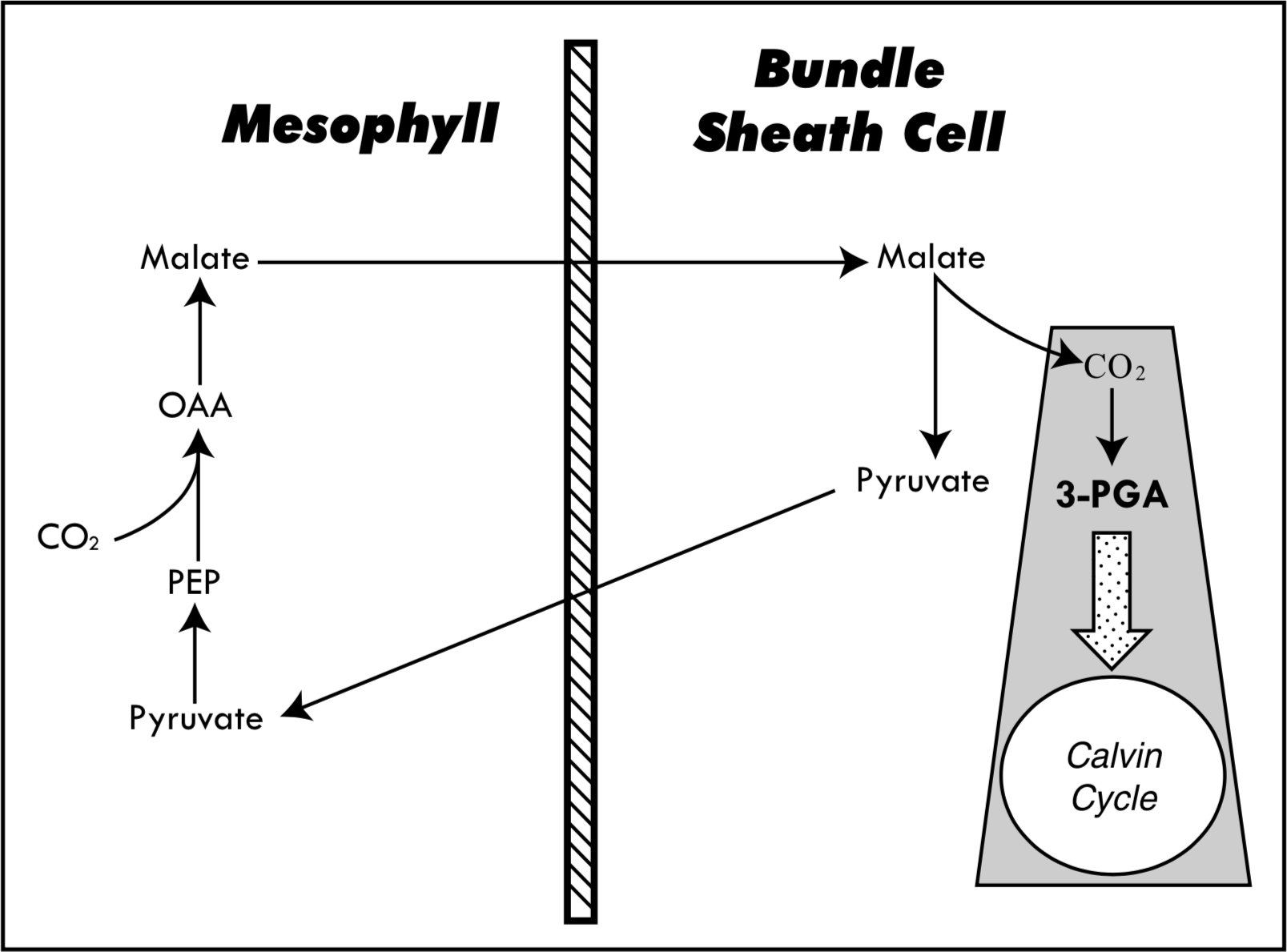
Although it may seem that this difference in biochemistry is sufficient to warrant the inference of direct divine design, the evidence is far from conclusive. There is no single C4 pathway; to date, three different subgroups have been described based primarily on the different enzymes used to liberate the CO2 from the four-carbon compounds transported to the BSC. The different subgroups use PEP carboxykinase, NADP-malic enzyme,and NAD-malic enzyme to catalyze this reaction. Furthermore, 23 different species in seven different genera and five different families have been identified as exhibiting traits intermediate between C3 and C4 photosynthesis (Monson and Moore 1989). These intermediate traits can be limited to simple anatomical changes in the cells surrounding the BSC, or may include biochemical alterations such as compartmentation of enzymes into the rudimentary BSC. All C3-C4 species possess a limited Kranz-like anatomy of specialized BSC that lack the thickened cell walls characteristic of true C4 BSC (Monson et al. 1984). Many of the BSC of C3-C4 species also concentrate organelles (mitochondria, chloroplasts, and peroxisomes) that contain enzymes critical to the C4 pathway. In Flaveria, a limited C4 pathway and enzymatic compartmentation has been observed in F. ramosissima (Monson et al. 1984). C3-C4 intermediates typically undergo photorespiration less than C3 species but more than C4 (Moore et al. 1987).
An irreducibly complex system is "composed of several well-matched, interacting parts that contribute to the basic function, wherein the removal of any one of the parts causes the system to effectively cease functioning" (Behe 1996). Thus, any system of interacting parts that continues to function in a limited fashion without one of the parts cannot be irreducibly complex by definition. The existence of true intermediates demonstrates that the C4 photosynthetic pathway is not irreducibly complex, for the C4 phenotype can continue to function in a limited fashion even when biochemical parts of the underlying mechanism are absent. Because of this reducible complexity in C4 photosynthesis, Behe's design inference cannot be applied, and the question of direct divine design remains (Behe 1996). Thus, if it were shown that C3, C4, and C3-C4 plants were members of the same holobaramin, the post-Creation origin of C4 photosynthesis would be implied by the majority of C3 species in Flaveriinae. Thus, Flaveria would be an excellent system to study the post-Creation origin of biochemical complexity.
RESULTS
Wise's baraminology matrix (1992) consists of a series of questions designed to allow researchers to detect phylogenetic discontinuity. Unfortunately, many of the questions in the matrix do not apply to the Flaveriinae. For instance, there is no mention of the Flaveriinae in the Scripture, nor could we find any reference to fossil Flaveriinae. Though the matrix cannot be used, the traditional baraminological practice of examining additive and subtractive evidence will be presented (Wise 1992, Robinson 1997, Robinson and Cavanaugh 1998a, Robinson and Cavanaugh 1998b ).
Additive Evidence. Hybridization between the members of Flaveriinae is extensive. In the wild, only F. floridana and F. linearis readily hybridize (Monson 1989). The remainder of the crosses summarized in Figure 2 are the result of artificial hybridization studies conducted by Powell (1978). These hybrids are very good evidence for the assignment of monobaramin status to each of the Flaveriinae genera. Of 59 Flaveria interspecific crosses reported by Powell, 40 were successful (Figure 2). Of the six possible interspecific crosses within Sartwellia, two were observed to be successful, and the two Haploëthes species were also successfully crossed. These crosses are strong indicators of the monobaraminic status of each genus of Flaveriinae.
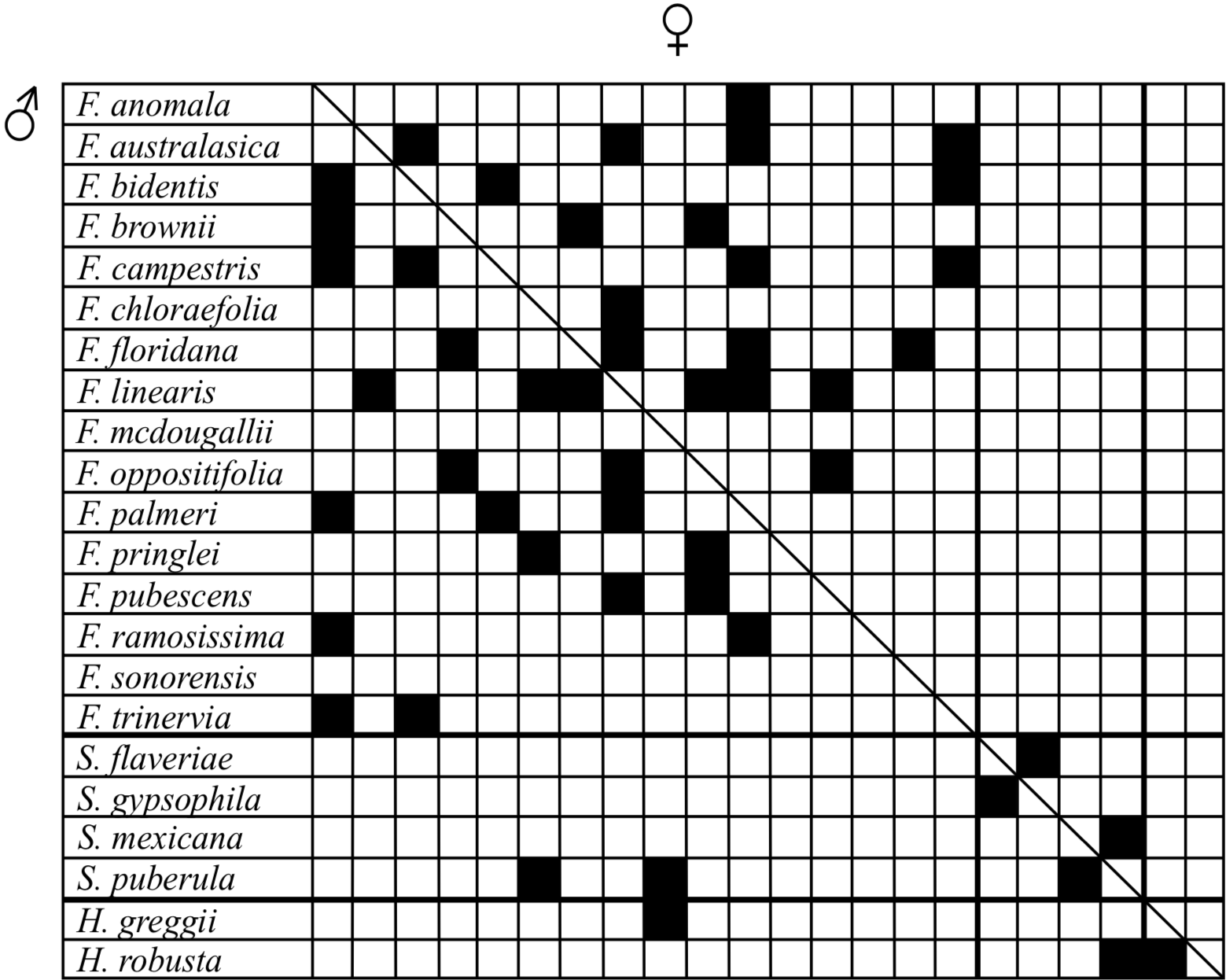
Hybridization within genus Flaveria reveals a significant pattern of hybridization success versus photosynthesis type (Figure 3). At the time of Powell's experiments in the 1970s, no C3-C4 intermediates had been conclusively identified, although they were suspected to exist. Nevertheless, examination of the hybridization data reported by Powell indicates a clear hybridization success bias between the different photosynthesis types. No cross between a C3 and C4 species was directly successful, indicating that perhaps the different photosynthesis types may themselves constitute monobaramins. The monobaramin status of genus Flaveria is not necessarily called into question by this data, if one assumes associative hybridization in baraminic assignments (Scherer 1993). For instance, F. pringlei (C3) crosses with F. oppositifolia (C3-C4) which crosses with F. linearis (C3-C4) which crosses with F. palmeri (C4). Thus, gene flow between C3 and C4 species can be achieved.
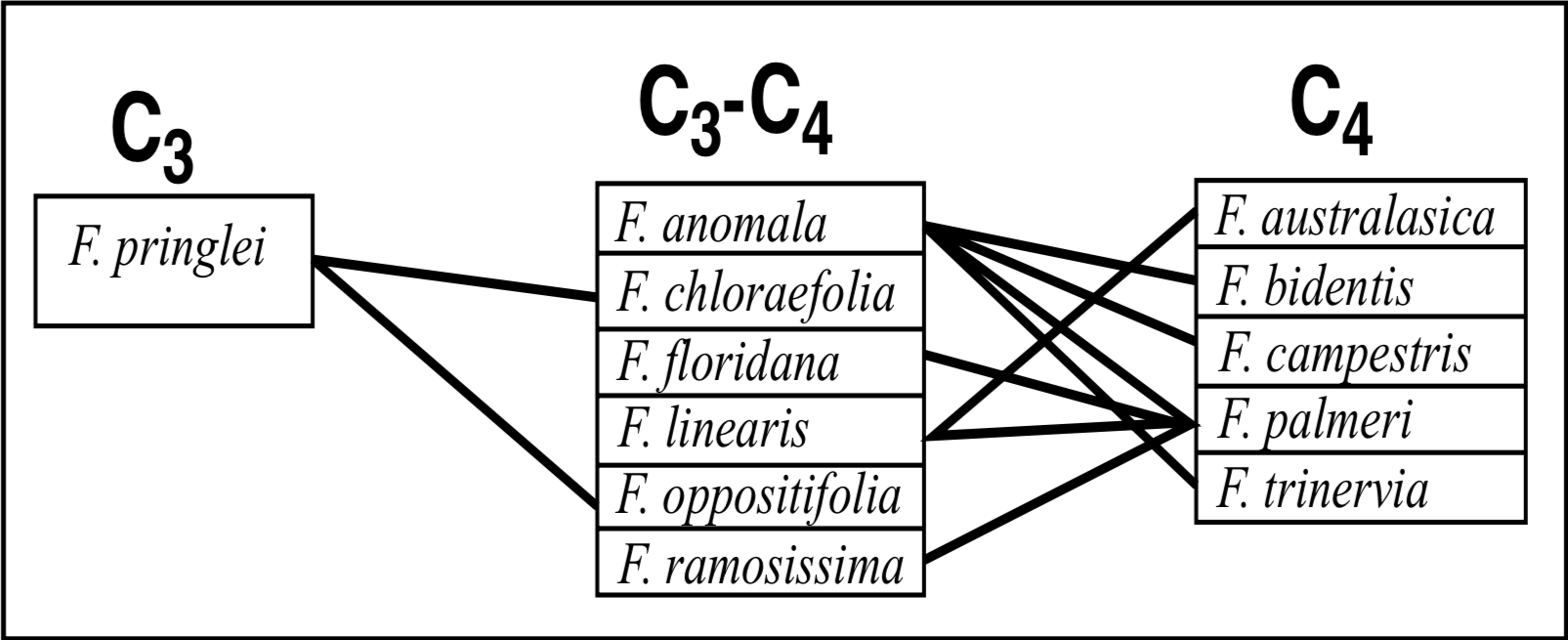
Powell also attempted more than 30 intergeneric crosses, but only four were successful. Significantly, these four intergenerics unite the species of all three genera, including the Flaveria outlier from Grand Canyon, F. mcdougallii, which hybridizes with no other Flaveria species. Again, by assuming associative hybridization, gene flow can be achieved between any species of Flaveria and F. mcdougallii via crosses with Sartwellia puberula (Figure 2). The sterile F1 intergenerics were described as "intermediate in morphology between the grossly different species involved" (Powell 1978). The success of the intergeneric crosses argues for the assignment of monobaramin status to the entire Flaveriinae subtribe, and the morphologically intermediate condition of these plants further supports this assignment (Wise 1992).
Ecologically, all species of Flaveriinae inhabit a similar niche, a dry, gypsiferous soil (Powell 1978, Turner 1971, Turner 1975). Wise assumed that the ecological criterion provides only weak evidence for phylogeny; however, in this instance, it may be significant that each species prefers not merely a dry but a gypsiferous soil. In combination with other evidence, this constitutes further evidence for the monobaraminic status of Flaveriinae.
The chromosome number for each Flaveriinae species is n=18. One specimen of F. campestris was reported as n=9; however, the author concluded it to be an isolated case of haploidy (Powell 1978). Specimens of F. pringlei have been reported to be n=36, but this is the only case of . polyploidy known for the subtribe (Powell 1978). It was therefore concluded that the fundamental chromosome number of Flaveriinae is x=18 (Powell 1978, Turner 1971, Turner 1975). This is further additive evidence for the monobaramin Flaveriinae; although by itself it is certainly inconclusive.
Limited DNA sequence information is available for 11 species of Flaveria. The gene for the H subunit of the glycine carboxylase system (gcsH) has been cloned and sequenced (Kopriva, Chu, and Bauwe 1996). Although this is a limited sample of the entire subtribe, it is sufficient to further establish the cobaraminic status of the C3, C4, and C3-C4 species. Of all the sequences, none are less than 93.2% identical. Most telling in this regard is the correlation of sequence similarity and hybridization potential (Figure 4). Three species (F. anomala, F. floridana, and F. linearis) can successfully hybridize with species that possess the most dissimilar sequences. Powell also noted that several Flaveria species have the ability to hybridize with very divergent Flaveria species, which he attributes to stronger reproductive barriers present in annual species than in perennial species (1978); of 40 successful interspecific crosses of Flaveria, only 5 are between an annual and perennial species. Further sequence analysis is necessary, particularly from specimens of Sartwellia and Haploësthes, before a full-sequence characterization of the Flaveriinae monobaramin can be made. Nevertheless, this sequence similarity certainly constitutes further additive evidence that the C3, C4, and C3-C4Flaveria species are cobaraminic.
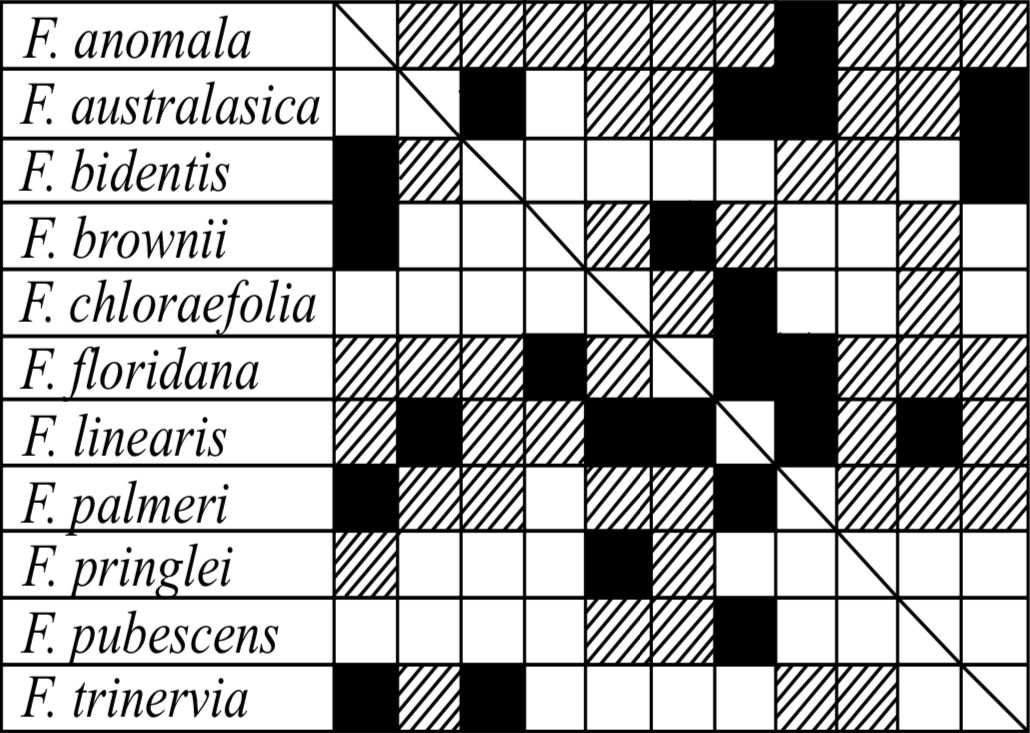
Subtractive Evidence. Unfortunately, subtractive evidence in the literature regarding subtribe Flaveriinae is scarce. No artificial hybridizations between a Flaveriinae species and a member of a separate subtribe have been reported; thus, the failure of hybridization gives no insights into reproductive isolation of the subtribe. Although the Flaveriinae possess 18 chromosomes universally, the predominant chromosome number for the tribe Helenieae is n=19, with other members of Flaveriinae sensu lato possessing 16 or 19 chromosomes (Lundberg 1996). The difference may be due to simple Robertsonian rearrangements, and in light of chromosomal rearrangements in mammals (Gibson 1984, 1986) and Arabidopsis chromosomes (Lin et al. 1999), it is unlikely that chromosome number will ever be a strong subtractive evidence. A phylogenetic analysis of Flaveriinae sensu lato has been conducted by Lundberg (1996). He found 12 equally parsimonious trees, all of which show a consistent clustering of the three genera Flaveria, Sartwellia, and Haploësthes apart from the other genera of Flaveriinae sensu lato. From the perspective of additive evidence, this is good reason to assign Flaveriinae the status of monobaramin, but Lundberg's dataset provides very poor subtractive evidence (see below).
No gcsH sequences from other members of tribe Helenieae are available for comparison to the known Flaveria sequences; however, a comprehensive analysis of the entire Asteraceae family has been done using the chloroplast gene NADH dehydrogenase (ndhF) (Kim and Jansen 1995). Included in this phylogeny were two members of Flaveriinae sensu lato, Jaumea carnosa and Flaveria ramosissima, and a species from subtribe Pectidinae (Tagetes erecta) of the same genus as outgroup species used by Lundberg (1996). The ndhF phylogeny consistently shows F.ramosissima more closely related to T. erecta than to J. carnosa (82% bootstrap value) (Kim and Jansen 1995). In contrast, the morphological analysis of Lundberg showed the genus Jaumea more closely related to Flaveria than either are to Tagetes (Lundberg 1996). Thus, the subtractive evidence for apobaraminic status of Flaveriinae is ambiguous at best.
Analysis of Pattern. Cavanaugh devised the ANOPA method specifically to resolve problems associated with reducing the multidimensional variation in morphological data to only a few dimensions without loss of data (Cavanaugh and Stemberg, in prep). A detailed discussion of ANOPA is beyond the scope of this paper, but a brief description of the method follows (for more details, consult Cavanaugh and Stemberg, in prep). In 1D ANOPA, a morphological centroid is calculated from the means of all numerically encoded characters. Next, distances (termed a0) from each taxon to the centroid are measured. In 2D ANOPA, an additional outgroup centroid is selected which is most distant from the initial centroid, and a "hyperline" is drawn between the two centroids [a hyperline is a one-dimensional line in multidimensional (>3D) space]. The hyperline serves as a line of reference to measure the distances t0 (along the hyperline) and d2 (from the hyperline) for each taxon. These multidimensional distances allow clustering patterns to be detected, including significantly similar species and significantly different species clusters. ANOPA is similar to discriminant analysis and principle components analysis in its ability to simplify highly complex, multidimensional datasets. Unlike discriminant analysis, ANOPA does not require any a priori categorization of the data points. ANOPA is also capable of recovering more information regarding the higher order pattern geometry without the loss of information as is incurred in PCA analysis. Of most significance to baraminologists is the fact that additive and subtractive evidence can be evaluated together in a single ANOPA analysis.
To apply ANOPA to the Flaveriinae, we used Lundberg's morphological data (1996). In addition to the sensu stricto Flaveriinae species discussed in this paper, Lundberg also included species from each genus of Flaveriinae sensu lato and four outgroup species from subtribe Pectidinae. The 1D ANOPA results (Figure 5) show no significant gaps between any of the species, including the Pectidinae. The 2D ANOPA results are very similar (Figure 6), with a slight gap between the C4 Flaveria species and the rest. Most significantly, the outgroup Pectidinae are intermingled with the members of Flaveriinae sensu lato, and Jaumea carnosa is very close to Sartwellia puberula. Based on these results, we would conclude that all of these species form a homogeneous cluster.
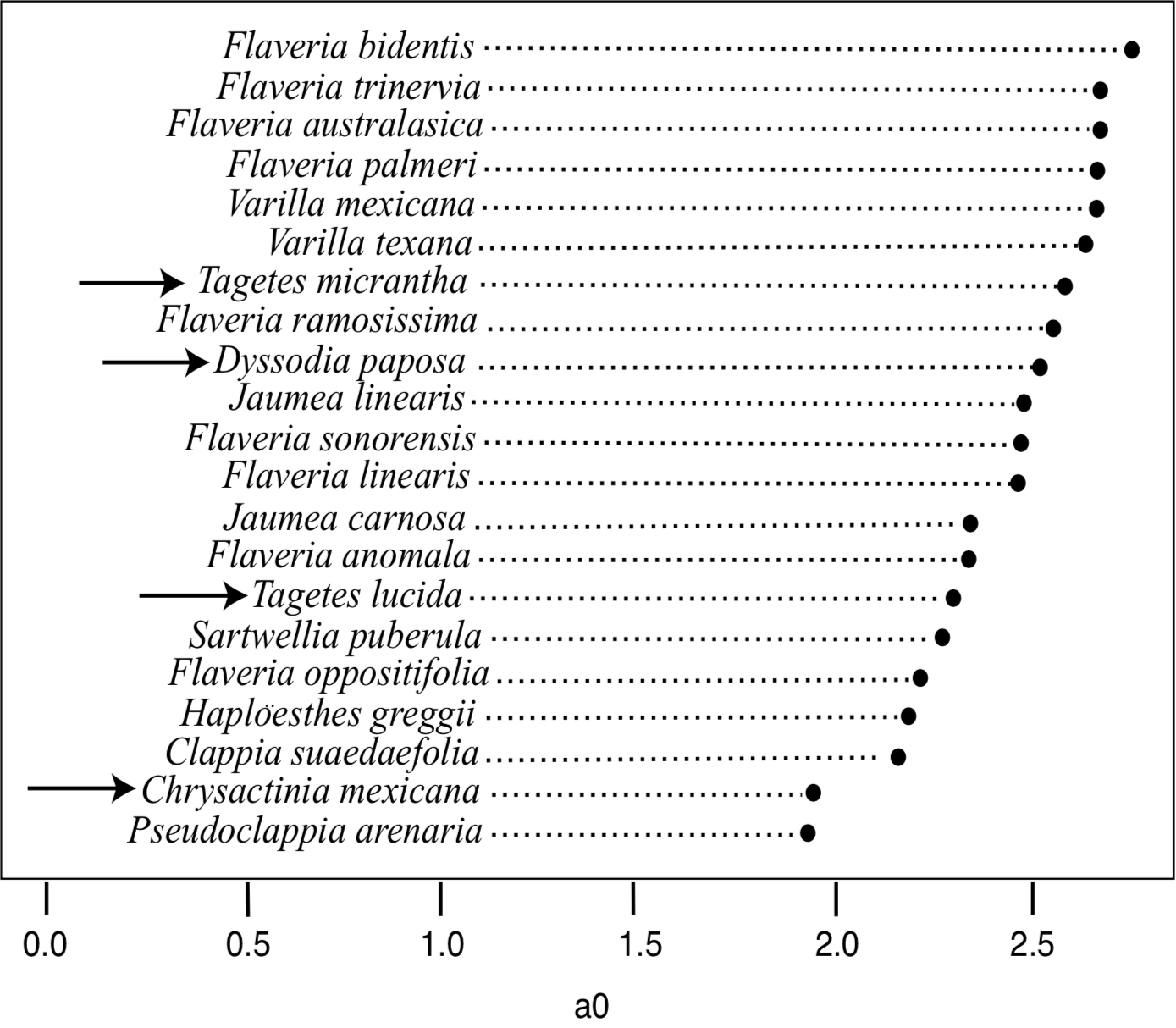
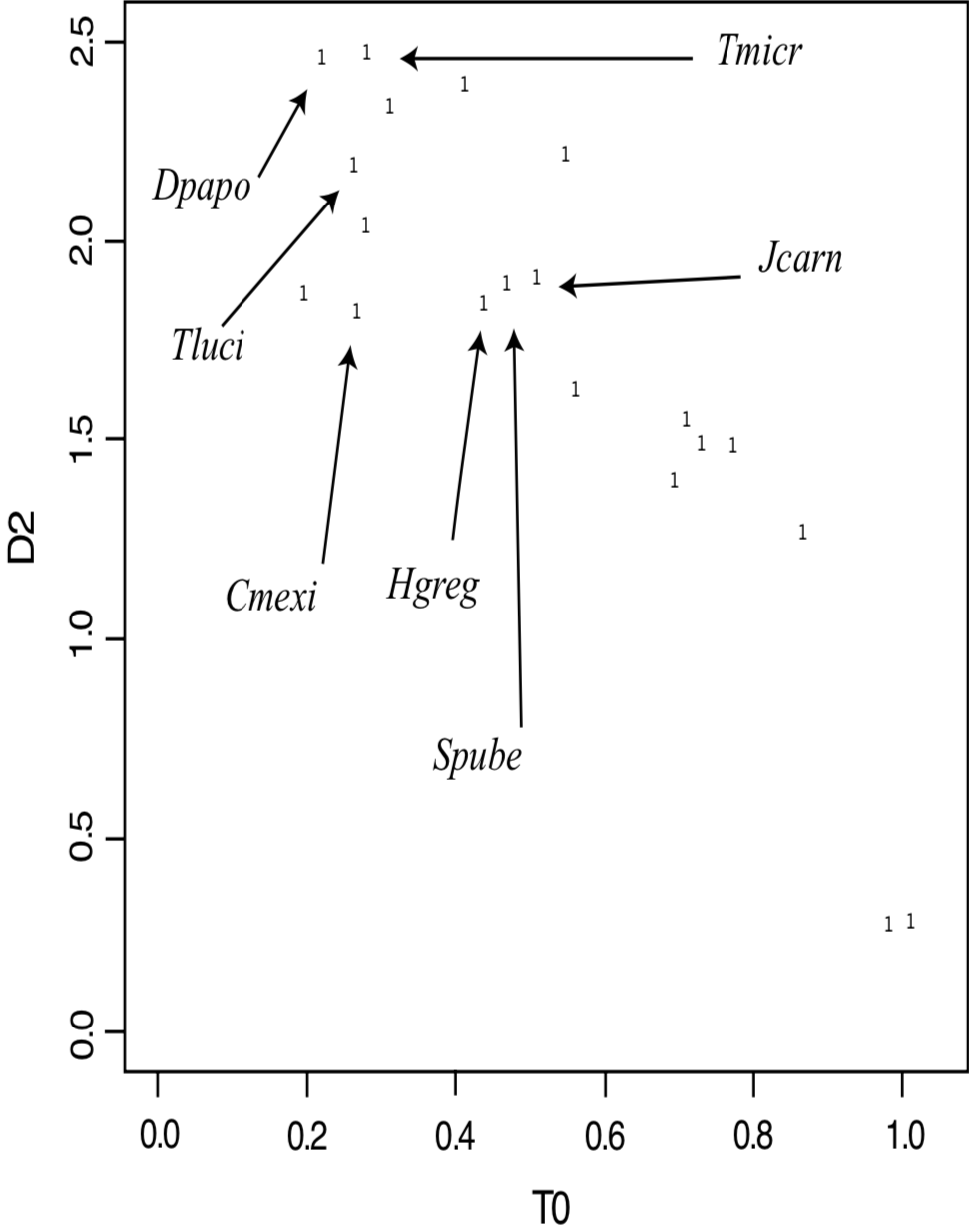
The baraminological interpretation of the ANOPA results is far from clear. Whereas the additive evidence strongly supports the monobaraminic status of Flaveriinae, the ANOPA results do not support apobaraminic status of the same group of species. This would suggest that Flaveriinae is probably a member of a larger holobaramin that includes at least the species of Flaveriinae sensu lato, and probably the species of Pectidinae as well. This would be consistent with the ndhF phylogeny that showed a close relationship between F. ramosissima and T. erecta. Unfortunately, the ANOPA results show no significant gaps between any of the species tested, thus it is not possible to establish the boundary of the apobaramin of which Flaveriinae is a part.
Summary. Although the additive evidence seems to support the monobaraminic status of Flaveriinae, as well as the monobaraminic status of Flaveria, Sartwellia, and Haploësthes individually, the subtractive evidence is far more ambiguous. Further research is necessary to clarify the apobaraminic status of Helenieae and Flaveriinae sensu lato, before an identification of the extant holobaramin can be made.
DISCUSSION
We initiated this study to address two questions: First, can the holobaramin be approximated as the family for nonvertebrates? Second, have C3, C4, and C3-C4 plant species descended from a common ancestor, thus implying the post-Creation emergence of biological complexity?
From our present analysis, we were unable to answer the first question definitively. We could not define any clear apobaraminic unit to which Flaveriinae belongs, allowing for the possibility that the entire family Asteraceae is a holobaramin. One observation in favor of this interpretation is the fossil record of the family Asteraceae. The family does not appear in the fossil record until the Oligocene and diversifies substantially by the Miocene (Bremer 1994, DeVore 2000). This Cenozoic appearance and diversification is similar to other post-Flood vertebrate baramins (Wise 1994, Wise 1995, Garner 1998, Wise 1999).
The second question regarding the origin of C4 photosynthesis has been answered quite clearly. All of our evidence supports the view that all species of the Flaveria genus are members of a single monobaramin and therefore share a common ancestor. Since C3 photosynthesis is the predominant type of photosynthesis for species of Flaveriinae and Flaveriinae sensu lato (Lundberg 1996), the most parsimonious interpretation is that the C4 species have developed from C3 ancestors. Based on this conclusion, the C4 photosynthetic pathway is a biochemical pathway that has emerged in Flaveria after Creation and quite possibly post-Flood (considering the fossil record of the Asteraceae mentioned above).
Although the sequence data are insufficient to aid in identifying the apobaramin to which the Flaveriinae belong, the sequences can assist in understanding the monobaraminic divisions within Flaveria itself and the evolution of Flaveria species. A phylogeny of the genus Flaveria rooted by a homologous sequence from Pisum sativum (the garden pea) is shown in Figure 7. The phylogeny shows three distinct groups of Flaveria sequences, corresponding to the C4, C3, and C3-C4 species groups. The phylogeny also shows that the C3-C4 group branches between the C3 and C4 groups, and this may be explained by one of two hypotheses. First, the occurrence of C4 and C3-C4 intermediates group on two different branches implies that the modern C3-C4 species are not true evolutionary intermediates between the modern C3 and C4 species but that extant C3-C4 intermediates are the living descendants of a true evolutionary intermediate population. Alternatively, one may also conclude that C4-like adaptations have arisen twice in the Flaveria, one producing the complete C4 syndrome and one producing only a partial C4 syndrome. To distinguish these explanations, we turn to the geography of the new world species of Flaveria. The distribution of the three photosynthetic types of Flaveria supports the hypothesis that the C3-C4 species are the descendents of the true evolutionary intermediates between the C3 and C4 Flaveria species. The C3Flaveria species and all species of Haploësthes and Sartwellia are restriced in range to Mexico and Texas, implying that the geographic origin of monobaramin Flaveriinae is in this region. The C3-C4 species of Flaveria are found in a restricted geographic band north of the C3 species (Figure 8). Further north still, the C4 species appear, from which they can then spread to a much broader range (Figure 8), including South America and the Australian F. australasica. It is likely that the C3-C4 intermediates are occupying a limited range because of their physiology, while the C4 plants are able to spread out much more readily because of their decreased photorespiration. Thus, when interpreted together, the molecular, morphological, biochemical, and geographic data all point to the evolutionary intermediacy of the ancestors of the modern C3-C4 species.
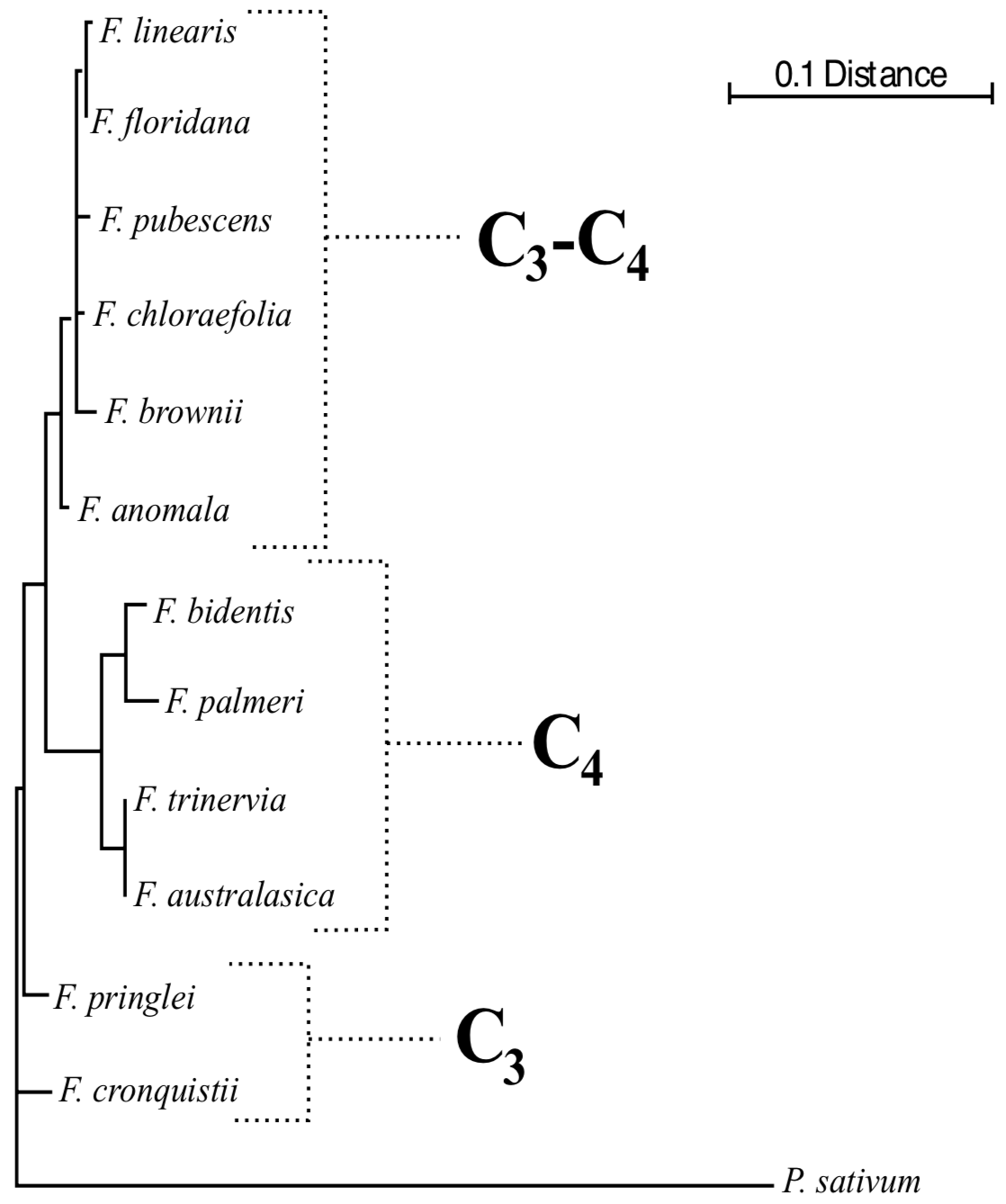
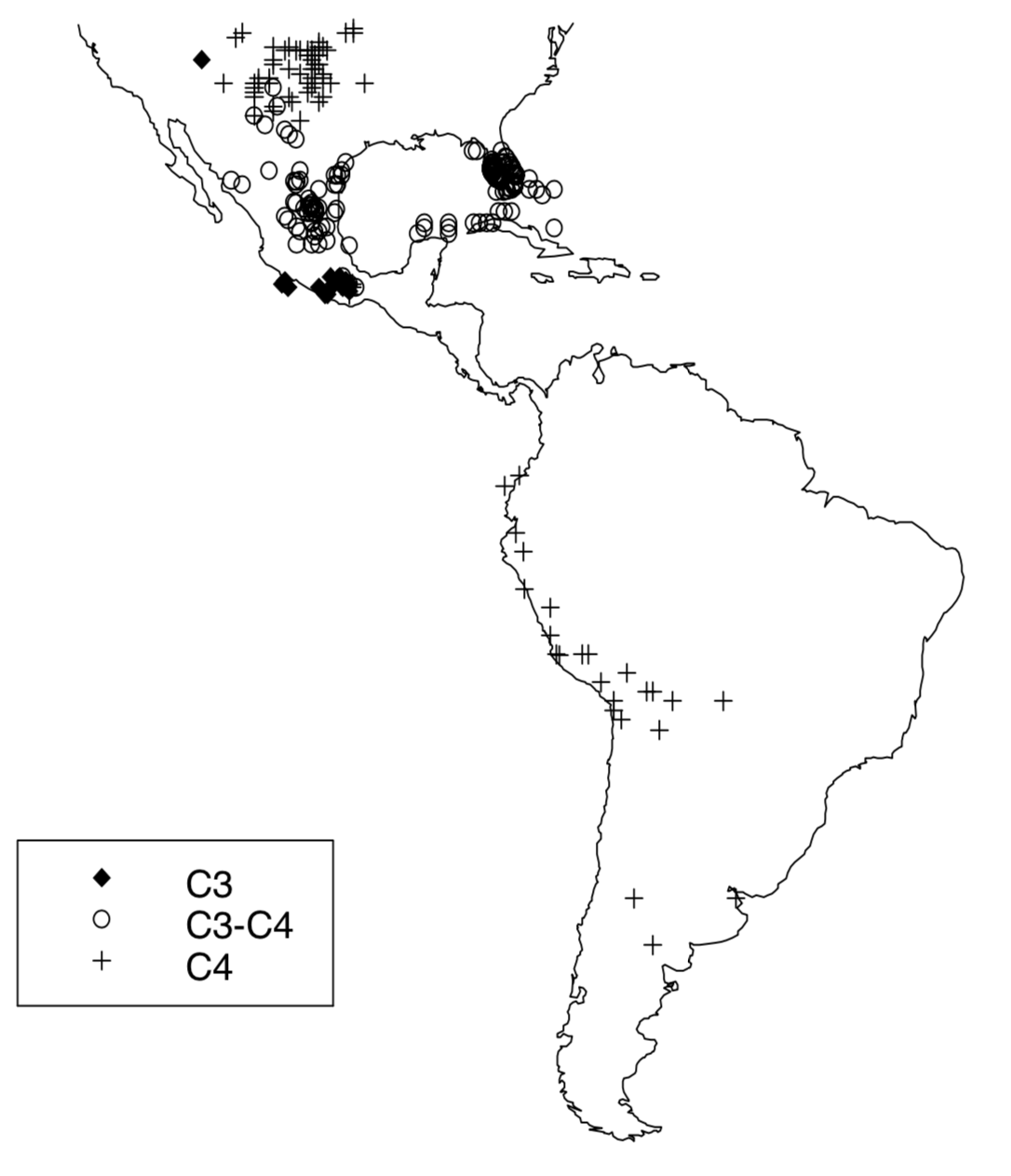
Because the modern C4Flaveria species descended from ancestral species that were C3, the C4 photosynthetic pathway must have arisen after the creation of this holobaramin. Thus, one explanation for the origin of C4 photosynthesis can be eliminated: direct creation by God. Superficially, the conclusion that C4 photosynthesis has arisen since creation seems to deny the basic creationist belief that no increase in biological complexity has occurred, that all evolution has been degenerative (Nelson 1967, Morris 1974, Williams 1976, Davis and Kenyon 1993, Sarfati 1997, Sarfati 1999). Eliminating direct creation does not, however, leave gradualistic, accidental evolution as the only alternative explanation. The emergence of C4 photosynthesis fits well with Wise's notion of diversification, a period of rapid speciation immediately following the Flood, which resulted in drastic changes within many holobaramins (Wise 1994, 1996). Recently, Wood has proposed a genetic model called the AGEing process that is capable of explaining numerous features of rapid post-Flood diversification (Wood, in prep). According to this model, intrabaraminic diversification occurs as a result of specially designed mobile DNA sequences called Altruistic Genetic Elements (AGEs). Hypothetically, AGEs are capable of activating or inactivating genetic potential already present in an organism's genome (Wood, in prep). Thus, C4 photosynthesis could be designed without being directly created in its complete form. God could have made the genes necessary for C4 photosynthesis in a latent state and allowed them to become activated at a later date by AGEs. The AGE-induced activation of latent genetic material explains the origin of C4 photosynthesis better than evolution for the following reasons:
- C4 photosynthesis is a complex characteristic resulting from the interaction of three levels of biological organization: tissue anatomy, cell structure, and biochemistry. All C4 plants possess specialized cells (BSC) in their leaves that house high concentrations of relevant organelles (Edwards and Walker 1983). Compartmentation also extends to important enzymes in the C4 pathway, especially rubisco. This coordinated evolution is difficult to explain with a simple mutation/selection scenario; however, if the information necessary to produce each of these traits was already present in the genomes of the holobaramin, AGEs could easily activate it during the post-Flood diversification.
- The previous reason would have much less force if it were not for the fact that C4 photosynthesis is not limited to a single holobaramin. C4 photosynthesis occurs in 16 different flowering plant families, two of which are monocots (Poaceae and Cyperaceae). While it is possible to imagine an evolutionary explanation of C4 photosynthesis once, it is very difficult to explain the parallel evolution of very similar traits at least 16 different times. Further, the grass family Poaceae appears to have developed C4 photosynthesis at least three times. All three types of C4 photosynthesis (NADP-ME, NAD-ME, and PEP-CK), each of which display different physical arrangements of chloroplasts in the BSC (Edwards and Walker 1983), are found among the grass species. The AGEing process can explain this type of parallelism both within and between baramins by either true transposition of genetic material or by activation of originally created genetic material common to both holobaramins (Wood, in prep).
- The fossil record of C4 plants consists of several grass specimens from the Pliocene/Upper Miocene (Thomasson et al. 1986, Nambudiri et al. 1978), but 13C/12C ratios in fossil herbivores indicate the presence of C4 grasses in the mid-Miocene (Morgan et al. 1994). A decrease in atmospheric CO2 levels in the Upper Miocene is often cited as the selectional pressure responsible for the radiation of the C4 grasses; however, if the C4 grasses are already present during the deposition of the mid-Miocene sediments, the Miocene drop in CO2 cannot have driven the initial emergence of the C4 pathway in the grasses (Morgan et al. 1994). Thus, the CO2 decrease merely serves to diversify a pathway that was already present in the grass population. The AGEing process predicts this type of phenotypic evolution prior to environmental selection for the AGE-induced trait (Wood, in prep).
If AGEing is indeed responsible for the expression of latent C4-specific genetic material in Flaveria, the various C3-C4 intermediates would represent species that have not yet had the full suite of C4 genes activated. Considering the potential for three apparently discrete levels of C4-like plants (anatomically C4, biochemically intermediate, and completely C4), this would imply a minimum of three different genetic changes necessary for the C4 trait to be fully expressed. Support for this theory is found in the genetics of C4 photosynthesis (Sheen 1999). In particular, C4-specific genes are known to occur in C3 species of Flaveria (Lipka et al. 1994). The primary differences between C4-specific genes in C3 and C4 plants is the promoter region which controls the expression of the gene (Sheen 1999). Further research into the physiology of C4 plants will no doubt shed new light on the source of these promoter differences in C3 and C4 species.
Regardless of mechanistic explanations, the presence of two very different kinds of photosynthesis in the same holobaramin highlights the elegant design that is so prevalent in the living world. Not only did God create organisms with what they needed at that time; He also provided an abundance of characteristics, which may or may not be immediately apparent, that would be necessary for survival in a world damaged by sin and ravaged by a worldwide Flood. From our narrow, utilitarian viewpoint, we may label a structure 'vestigial' or a strand of DNA 'junk,' but given the proper circumstances, these useless features may prove their value after all. The notion of AGE-activated latent genetic material could become a powerful explanation of apparently useless features of living organisms as simply the unexpressed abundance of a benevolent Creator.
ACKNOWLEDGMENTS
A very early draft of this paper was presented at a meeting of the Baraminology Study Group held at Bryan College, June 27, 1997. We thank Ashley Robinson, Kurt Wise, and our anonymous reviewers for their critical reading of various drafts of this work. Many thanks to Stephanie Mace for assistance in preparing Figure 8.
METHODS
ANOPA. The cladistic data matrix of Lundberg (1996) was used for ANOPA analysis. Because ANOPA requires all data to be coded numerically, unknown character states were recoded as 0's with subsequent states increased by 1. Thus, a character state in Lundberg coded as ?, 0, and 1 is here recoded as 0, 1, and 2, respectively. A complete matrix of the transformed data is available from the authors on request. ANOPA was performed using standard methodology as described by Cavanaugh and Sternberg (in prep).
Molecular Phylogenetics. The sequence phylogeny was constructed from gcsH sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov) for the following species: F. anomala (Z37524), F. bidentis (Z37517), F. chloraefolia (Z37520), F. cronquistii sequence A from Kopriva et al. (1996) (Z25854), F. floridana (Z37528), F. linearis (Z37521), F. palmeri (Z37529), F. pringlei sequence A from Kopriva et al (1996) (Z25855), F. pubescens (Z37530), F. trinervia (Z37523), and Pisum sativum (J05164). We omitted loci B and C from the C3 plants for simplicity and because the source of the F. pringlei gcsHB is questionable (see discussion in Kopriva et al. 1996). Sequences were aligned with CLUSTAL W v. 1.75 (Thompson et al. 1994) using default parameters. After trimming end-gaps, the final alignment consisted of 359 positions. Distances were inferred using the DNADIST program of the PHYLIP package (Felsenstein 1993). The phylogeny was constructed using the Fitch-Margoliash method (Fitch and Margoliash 1967) as implemented in the FITCH program of the PHYLIP package.
LITERATURE CITED
Behe MJ. 1996. Darwin's black box. NY: The Free Press.
Bremer K. 1994. Asteraceae: cladistics & classification. Portland, OR: Timber Press.
Cavanaugh DP, Sternberg RV. In preparation. Analysis of morphological constraints using ANOPA, a pattern recognition and multivariate statistical method: a case study involving centrarchid fishes.
Cronquist A. 1981. An integrated system of classification of flowering plants. NY: Columbia University Press.
Davis P, Kenyon DH. 1993. Of pandas and people, 2nd ed. Dallas, TX: Haughton Publishing Company.
DeVore ML. 2000. The role of paleobotanical and geological data in biogeographical studies of groups with limited fossil records: an example using Asteraceae. Geological Society of America Abstracts with Programs 32(7):A194.
Edwards G, Walker D. 1983. C3, C4: mechanisms, and cellular and environmental regulation, of photosynthesis. Los Angeles, CA: University of California Press.
Felsenstein J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Seattle, WA: Department of Genetics, University of Washington.
Fitch WM, Margoliash E. 1967. Construction of phylogenetic trees. Science 155:279-284.
Garner P. 1998. It's a horse, of course! A creationist view of phylogenetic change in the equid family. Origins (UK) 25:13-23.
Gibson LJ. 1984. Chromosomal changes in mammalian speciation: a literature review. Origins 11:67-89.
Gibson LJ. 1986. A creationist view of chromosome banding and evolution. Origins 13:9-35.
Jones AJ. 1972. Boundaries of the min: an analysis of the mosaic lists of clean and unclean animals. Creation Research Society Quarterly 9:114-123.
Karis PO, Ryding O. 1994. Tribe Helenieae. In Bremer K, editor. Asteraceae: cladistics & classification. Portland, OR: Timber Press.
Kim K-J, Jansen RK. 1995. ndhF sequence evolution and the major clades in the sunflower family. Proceedings of the National Academy of Sciences (USA) 92:10379-10383.
Kopriva S, Chu C-C, Bauwe H. 1996. Molecular phylogeny of Flaveria as deduced from the analysis of nucleotide sequences encoding the H-protein of the glycine cleavage system. Plant, Cell and Environment 19:1028-1036.
Lin X, Kaul S, Rounsley S, Shea TP, Benito M-I, Town CD, Fujii CY, Mason T, et al. 1999. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402:761-768.
Lipka B, Steinmüller K, Rosche E, Börsch D, Westhoff P. 1994. The C3 plant Flaveria pringlei contains a plastidic NADP-malic enzyme which is orthologous to the C4 isoform of the C4 plant F. trinervia. Plant Molecular Biology 26:1775-1783.
Lundberg J. 1996. Phylogeny of subtribe Flaveriinae (Asteraceae: Helenieae). Unpublished graduate thesis, Uppsala University.
Marsh FL. 1976. Variation and fixity in nature. Mountain View, CA: Pacific Press Publishing Assn.
Monson RK. 1989. On the evolutionary pathways resulting in C4 photosynthesis and Crassulacean acid metabolism (CAM). Advances in Ecological Research 19:57-110.
Monson RK, Moore BD. 1989. On the significance of C3-C4 intermediate photosynthesis to the evolution of C4 photosynthesis. Plant, Cell and Environment 12:689-699.
Monson RK, Edwards GE, Ku MSB. 1984. C3-C4 intermediate photosynthesis in plants. BioScience 34:563-574.
Moore BD, Ku MSB, Edwards GE. 1987. C4 photosynthesis and light-independent accumulation of inorganic carbon in leaves of C3-C4 and C4Flaveria species. Australian Journal of Plant Physiology 14:657-668.
Morgan ME, Kingston JD, Marino BD. 1994. Carbon isotopic evidence for the emergence of C4 plants in the Neogene from Pakistan and Kenya. Nature 367:162-165.
Morris HM, editor. 1974. Scientific creationism. San Diego, CA: Creation-Life Publishers.
Nambudiri EMV, Tidwell WD, Smith BN, Hebbert NP. 1978. A C4 plant from the Pliocene. Nature 276:816-817.
Nelson BC. 1967. After its kind. Minneapolis, MN: Bethany Fellowship.
Powell AM. 1972. Artificial hybridizations in the subtribe Peritylanae (Compositae-Helenieae). American Journal of Botany 59:760-768.
Powell AM. 1978. Systematics of Flaveria (Flaveriinae-Asteraceae). Annals of the Missouri Botanical Garden 65:590-636.
Robinson DA. 1997. A mitochondrial DNA analysis of the testudine apobaramin. Creation Research Society Quarterly 33:262-272.
Robinson DA, Cavanaugh DP. 1998a. A quantitative approach to baraminology with examples from the catarrhine primates. Creation Research Society Quarterly 34:196-208.
Robinson DA, Cavanaugh DP. 1998b. Evidence for a holobaraminic origin of the cats. Creation Research Society Quarterly 35:2-14.
Sarfati JD. 1997. Blood types and their origin. Creation Ex Nihilo Technical Journal 11:31-32.
Sarfati J. 1999. Refuting evolution. Green Forest, AR: Master Books.
Scherer S. 1993. Basic types of life. In Scherer S, editor. Typen des Lebens. Berlin: Pascal-Verlag.
Sheen J. 1999. C4 gene expression. Annual Review of Plant Physiology and Plant Molecular Biology 50:187-217.
Siegler HR. 1978. A creationist's taxonomy. Creation Research Society Quarterly 15:36-38,11.
Thomasson JR, Nelson ME, Zakrzewski RJ. 1986. A fossil grass (Gramineae: Chloridoideae) from the Miocene with Kranz anatomy. Science 233:876-878.
Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22:4673-4680.
Turner BL. 1971. Taxonomy of Sartwellia (Compositae Helenieae). SIDA 4:265-273.
Turner BL. 1975. Taxonomy of Haploësthes (Asteraceae-Senecioneae). Wrightia 5:108-119.
Williams EL. 1976. A creation model for natural processes. Creation Research Society Quarterly 13:34-37.
Wise KP. 1992. Practical baraminology. Creation Ex Nihilo Technical Journal 6:122-137.
Wise KP .1994. Australopithecus ramidus and the fossil record. Creation Ex Nihilo Technical Journal 8:160-165.
Wise KP. 1995. Towards a creationist understanding of 'transitional forms.' Creation Ex Nihilo Technical Journal 9:216-222.
Wise KP. 1996. North American Paleontology Convention 96. Creation Ex Nihilo Technical Journal 10:315-321.
Wise KP. 1999. The Camelidae fossil record. Presented at Baraminology 99, August 5-7, 1999, Liberty University, Lynchburg, VA.
Wood TC. In preparation. The AGEing process: Post-Flood intrabaraminic diversification caused by Altruistic Genetic Elements (AGEs).
Woodmorappe J. 1996. Noah's ark: a feasibility study. Santee, CA: Institute for Creation Research.