©Copyright 2018 GEOSCIENCE RESEARCH INSTITUTE
11060 Campus Street • Loma Linda, California 92350 • 909-558-4548

Imagine walking down the beach and coming across the words “Romeo loves Juliet” written in the sand. Most of us have experienced something like this and would not be surprised, but most people would be surprised to find the entire text of William Shakespeare’s play Romeo and Juliet written in sand. Why is this? The obvious reason is that sand is the wrong material for large writing projects. Sand grains move about easily and text written in it is quickly obliterated (Figure 1). Before the first act was completely written in sand, the beginning may be gone if the wind is blowing or waves wash over it.

On the other hand, it is not surprising to find Romeo and Juliet written out on paper. Because it is compact and lasts for many years, paper is an excellent material for storing information. We could write information on other materials – over the course of history, humans have employed everything from rocks to parchment made from sheep skins as information storage media – but cheap abundant long-lasting paper has proven to be one of the best media ever invented for storing information.
DNA – The stable genetic information storage medium
Inside cells, we find an information storage medium that, like paper, appears to be just the right medium for the information it stores; we call it Deoxyribonucleic Acid or DNA. One reason DNA is an excellent storage medium for genetic data is its amazing chemical stability. DNA can last for thousands of years after an organism has died, and this has allowed us to read the DNA of long extinct organisms ranging from our Neanderthal relatives[1] to woolly mammoths.[2] Chemical stability is essential to the function of DNA. If it were unstable, the information that it encodes would be quickly degraded, like information written in sand.
DNA – An efficient storage medium
Our own DNA illustrates another essential attribute of DNA. The human genome comprises all our DNA packaged into the 23 human chromosomes, two copies of which are found in most cells,[3] as well as DNA within mitochondria. DNA stores information far more efficiently than printed words on paper. For example, human mitochondrial DNA is only about 5.6 μm long–about one hundredth the diameter of a typical period at the end of a sentence–but printed out as letters on paper, its DNA sequence takes up about 6 pages. Similarly, the entire human genome is about a meter long but requires around a million pages to print out! Clearly a million pages will not fit inside a cell, but all that information will fit inside the tiny nucleus of a cell if it is encoded in DNA. The ability of DNA to store information in a tiny space makes it the most efficient known information storage medium in terms of information density.
DNA – An accurately copied medium
The double helical structure of DNA also contributes in an amazing way to its function as an information storage medium. When most cells divide, it is necessary to make a complete copy of the DNA in the “mother” cell so that each “daughter” cell gets one complete copy of the mother cell’s DNA. Information is encoded in DNA using sequences of chemicals called nucleotide bases. These bases are analogous to the letters we use to spell out information and a complete copy of the human genome contains about 3 billion of them. When human cells divide, the challenge is to accurately copy 6 billion bases because each mother cell contains two copies of the human genome.
Humans start out as a single fertilized egg cell, which divides until a person reaches the adult number of cells, which is around 37 trillion.[4] As adults, our cells continue to divide and grow as we lose old cells, so millions of cells in our bodies divide every day and thus millions of copies of the 6 billion bases in each cell’s DNA must be accurately made. While the number of DNA replications and the 3 billion base length of the human genome may seem impressive, this is not exceptional. Many other organisms, ranging from maize to some salamanders, have significantly larger genomes. The largest animal genome reported is for the marbled lungfish, Protopterus aethiopicus, with 139 billion bases in its genome, but organisms with even larger genomes are known.[5]
When James Watson and Francis Crick published their seminal 1953 paper[6] revealing the double helical structure of DNA to the world, they immediately noted that the structure suggests a mechanism for accurate DNA replication:
It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.
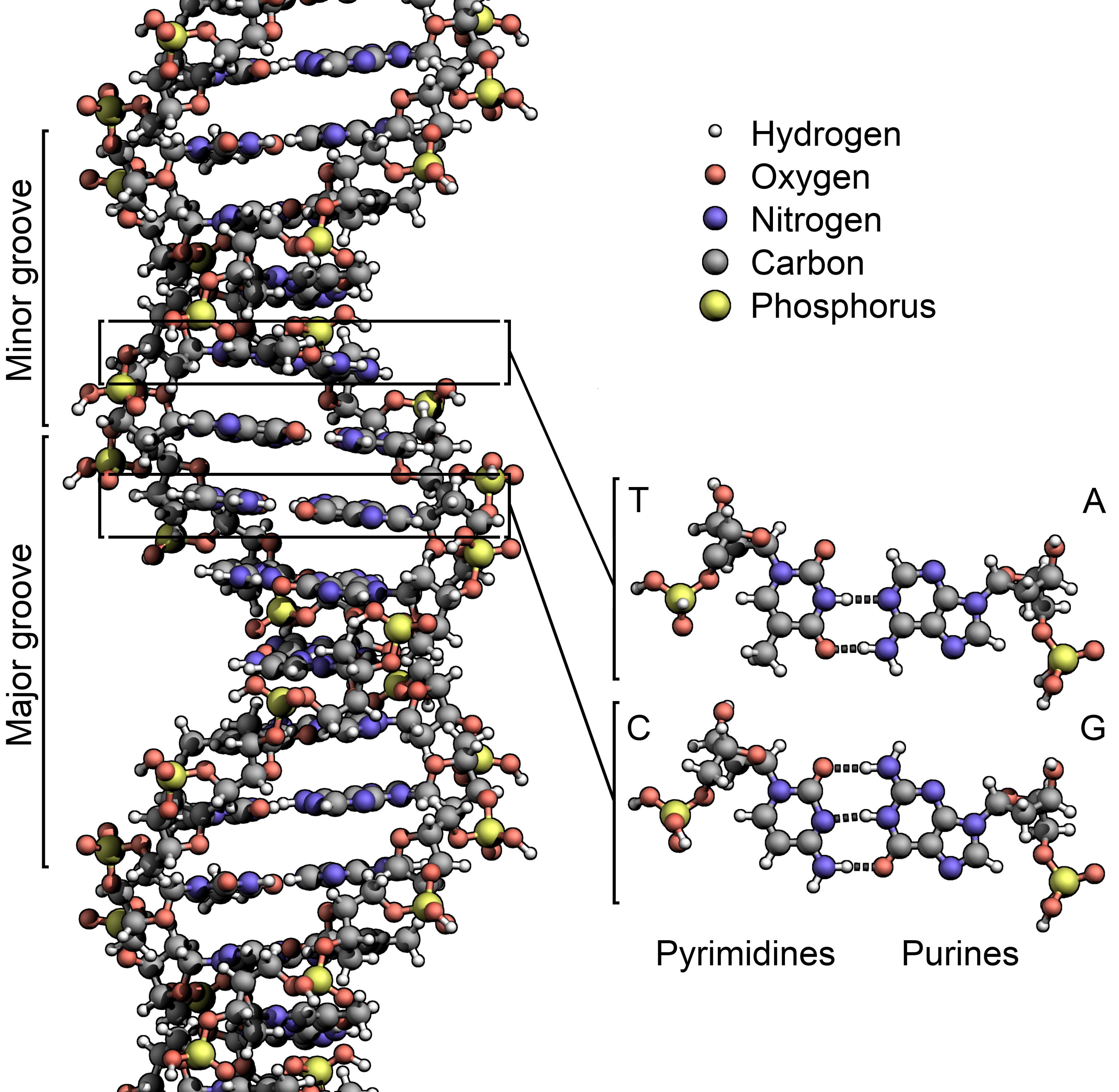
The pairing Watson and Crick are referring to involves the chemical base “letters” of the DNA alphabet already mentioned. These flat molecules are attached to the two strands of the double helix. As the two strands of the double helix wind around each other, the bases point into the center and interact with one another in very specific ways. One class of bases, the purine bases Adenine (A) and Guanine (G), is relatively large; while the other class, the pyrimidine bases Thymine (T) and Cytosine (C), are smaller. In addition, charges on the surface of these bases are distributed differently. Because of this and the geometry of DNA with the bases pointing toward each other, Adenine (A) on one strand of the double helix always pairs up with Thymine (T) on the opposite strand and vice versa. The same is true for Guanine (G) and Cytosine (C). Thus, if you have an A on one strand of the double helix, you can know that there is a T on the opposite strand, and if you have a C on one strand, you can know that there is G on the opposite strand.
If the double helix is unzipped to make individual strands of DNA, each strand contains a negative copy of the opposite strand. When DNA is replicated, the DNA double helix is unzipped and each strand serves as the template for making an exact copy of the other strand. Obviously there is complex cellular machinery that makes this happen, but if one strand has the sequence AGTCCGC, then the opposite strand can be reconstructed exactly from it as TCAGGCG.[7] Thus it is that the structure of DNA contributes to the amazing way it can be replicated with almost inconceivable speed and accuracy.
The origin of DNA as the genetic material of organisms
The more we learn about DNA, the better suited it appears to its role as an information storage medium inside cells. We have talked about three ideal characteristics that it exhibits: 1) It is chemically stable and thus does not rapidly degrade the information stored in it, 2) DNA is amazingly information dense, storing massive quantities of information in an incredibly small volume; 3) The structure of DNA contributes to its ability to be accurately copied at amazing speed, which is necessary when cells are dividing to make new cells. There are other characteristics that DNA exhibits that make it ideal, but discussing them gets progressively more technical and these three are sufficient to illustrate the point. There may be some other materials that could do some of what DNA does, but none are known that fulfill all the requirements of a genetic material as well as DNA does.
This raises the question of how organisms ended up using DNA rather than some inferior molecule to store genetic information. For the open-minded there are at least two possibilities, either DNA was chosen as the genetic material by someone who knew what they were doing, or DNA was chosen as the genetic material by something that didn’t know what it was doing. The latter position is the materialistic belief of Darwinism. How might this have worked? Given the amazing fit between the properties of DNA and its function, and the diverse molecules made by organisms, it is hard to imagine nature arrived at this solution on its first attempt. Something had to be the initial genetic material and then various other solutions to the need for a genetic material were tried until natural selection settled on the essentially ideal solution of DNA.
There is a significant problem with this Darwinian scenario. Darwin pointed out that for his mechanism to work, it must proceed by “by numerous, successive, slight modifications.”[8] However, changing genetic materials, even with relatively small changes to the chemicals involved, is not possibly a slight modification from the perspective of the organism. An analogy might be trying to swap out the hard disk of a computer for paper, or information chiseled into rock. Information is stored in the form of different magnetic states on hard drives, the equipment to retrieve that information must match the medium it is stored in. Thus, the heads that read changes in magnetic state on a hard disk are incapable of reading ink on paper or letters carved into stone. The same is true of the equipment used by cells to read information from the genetic material of cells. Today we can study the machinery found inside cells and see that we can’t substitute a different kind of molecule for DNA. Molecules like polysaccharides, triglycerides or proteins – all of which are naturally made inside cells – are not particularly good information storage media, but even if they were, they are not read by the machinery that reads DNA.
We can also see that devices capable of reading different media are a complex engineering challenge, but creating a device flexible enough to read multiple media is orders of magnitude more difficult. This is why when the transition was made from optical drives to USB drives no attempt was made to create systems that read both; they are read by different devices inside computers. To go with the materialistic scenario involving multiple genetic materials, one needs mechanisms for reading the stored information in different media that somehow anticipate the need to read it before it is being used. In reality, switching genetic materials, even switching between fairly closely related chemicals, requires a coordinated change in several molecular machines inside cells, something that seems quite remarkable in any system, let alone a system that is unguided in any way.
The alternative theory, that DNA was chosen as the genetic material by someone who knew what they were doing, can be compared with our normal experience. One of the first things engineers look at when presented with an engineering problem is the materials available and a decision is made about which materials will best serve the purposes of the problem they seek to address. For example, when designing a car engine, an engineer needs to use materials that can withstand the heat and mechanical strain inside the engine. Water is not the right material for an engine block, neither is wood or concrete; certain metals work very well for this purpose, so these metals are the materials most commonly used. Engineers know the specifications of the project being worked on and the specifications of the materials available. They then match the material exhibiting the necessary specifications with the project at hand. This observation fits well with our understanding of DNA as the genetic material; it is well explained within the paradigm of a wise Designer who chose an ideal material for the purpose of storing genetic information within the organisms He created. Satisfied with this choice, and many others, He could thus pronounce at the completion of His creation that it was “very good.”[9]
There is clearly an argument to be made for design in living things based on the choice of DNA as the genetic material, but what about the information encoded in DNA? Christians believe God created living things and is the ultimate source of information in genomes. In contrast, the most common alternative theory of origins–materialistic Darwinism–attributes genomic information to chance mutations and natural laws, particularly natural selection. Let’s look at two attributes of genomes that shed light on their origin and ultimately the origin of life.
Is much of the information coded in DNA actually junk?
Our understanding of genomes was held back by the rush to declare most of many genomes “junk.”[10] Susumu Ohno, who coined the term “junk DNA,” elegantly expressed this Darwinian viewpoint:
"[T]he earth is strewn with fossil remains of extinct species; is it a wonder that our genome too is filled with the remains of extinct genes?"[11]
Much DNA does not code for proteins, and many biologists initially assumed that these non-coding DNA sequences therefore lacked function. The logic boiled down to, “if we don’t know what it does, it must do nothing.” However, much data now demonstrate that this logic leads to a false conclusion: non-coding DNA often exhibits important functions. This understanding has revolutionized how biologists view genomes. Instead of vast deserts with occasional oases of functional information, genomes now seem more like rainforests of information with a dazzling array of genes, control mechanisms and logic circuits.
Genes are smarter than we thought
According to the “one gene, one enzyme” model proposed by Beadle and Tatum (for which they won a Nobel Prize), each gene codes for a single protein; but now everything has changed. Current estimates indicate that humans have less than 25,000 genes, but produce more than 100,000 proteins;[12] thus at least some genes must be capable of producing multiple proteins.
How is this achieved? Let’s illustrate with the human Paired-like homeodomain transcription factor 2 gene (also called Pituitary Homeobox 2 and abbreviated Pitx2), which shows how RNA processing creates several different proteins from one gene. The PITX2 protein (Figure 3) plays a role in development of the head and eyes, among other things.[13] Pitx2 includes six segments called exons that code for parts of the protein. These are separated by five introns. When the DNA coding this gene is copied to RNA for use by the cell’s protein manufacturing machinery, it can be processed in different ways to code for different proteins. By joining exons 1, 2, 5 and 6, mRNA for a version of PITX2 called “Isoform A”, or PITX2A is made. Joining exons 1,2,3,5 and 6 makes mRNA for PITX2B and exons 4, 5 and 6 form the mRNA for PITX2C. Other mechanisms make even more forms of PITX2.[14] Cells need to make the right “version” of PITX2 in the right place at the right time if the organism they are part of is to develop normally.
How does the cell “know” when to make one form of a protein but not others? This brings us back to some of what was once dismissed as “junk” DNA and indicates why genomes are now understood to be far more dynamic than initially imagined. The complex systems controlling exon splicing appear to involve sequences occupying at least one third of the human genome,[15] far exceeding the 3 % of the human genome thought to be functional a few years ago. The picture of DNA that is emerging shows it does in fact store vast quantities of information, much of which was missed by the first scientists to study it. Much more is probably yet to be discovered.
Conclusions
There is beauty, complexity, elegance and efficiency in the choice of DNA as the medium for storing genetic information, and the information itself is amazing. Some of what we know about genomes fits well with the thesis of common ancestry and Darwinian evolution, but when taken as a whole, the evidence is more consistent with the Biblical worldview in which a Designer, God, accounts for the choice of DNA as the genetic material, the enormous quantities of information stored in it and the elegant arrangement of that information in genomes.
When the Creator God wrote the 10 Commandments,[16] He did so in the durable medium of stone; but when He wrote the sins of the self-righteous scribes and Pharisees, He wrote in dust, where His words would be quickly obliterated.[17] The God of the Bible demonstrated His ability to choose appropriate media in which to record information, it should not be surprising to find that in living things He chose a medium as well suited to the task of recording genetic information as DNA, or that the information stored in DNA is elegant and amazing. Every person, every creature, every plant–indeed every living organism–is an exquisite repository of genetic information encoded in DNA, information far more exquisite and deeply meaningful than anything written by William Shakespeare or any other human author. Christians have good reason to believe that we are “fearfully and wonderfully made”[18] and that God, who created us along with all other things, is worthy of our worship.
References
[1] Green RE, Krause J, Briggs AW, et al. 2010. A Draft Sequence of the Neandertal Genome. Science 328(5979):710-722. DOI: 10.1126/science.1188021
[2] Miller W, Drautz DI, Ratan A, et al. 2008. Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456:387-390. doi:10.1038/nature07446
[3] We get a complete set of human chromosomes from each parent; 23 from our mother and 23 from our father for a total of 46. Most human cells carry all 46 chromosomes in their nucleus.
[4] Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-Amodio S, Strippoli P, Canaider S. 2013. An estimation of the number of cells in the human body. Annals of Human Biology. 40(6):463-71. doi: 10.3109/03014460.2013.807878.
[5] Pellicer J, Fay MF, Leitch AJ. 2010. The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society 164(1):10–15. DOI: 10.1111/j.1095-8339.2010.01072.x
[6] Watson JD, Crick FHC. 1953. A structure for deoxyribose nucleic acid. Nature 171:737-738.
[7] Note that because of the conventions usually used by molecular biologists, these sequences would not be written exactly like this.
[8] Darwin, C. R. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray. 1st edition, 1st issue. P 189.
[9] Genesis 1:31
[10] Makalowski, W. 2003. Not junk after all. Science 300:5623.
[11] Ohno S. 1972. So much "junk" DNA in our genome. Brookhaven symposia in biology. P. 366-70 in Evolution of genetic systems (H. H. Smith, ed.). Gordon and Breach, New York.
[12] Clamp, M., B. Fry, M. Kamal, X. Xie, J. Cuff, M. F. Lin, M. Kellis, K. Lindblad-Toh, and E. S. Lander. 2007. Distinguishing protein-coding and noncoding genes in the human genome. Proceedings of the National Academy of Sciences USA 104:19428–19433.
[13] Gage PJ, Suh H, Camper SA. 1999. The bicoid-related Pitx gene family in development. Mammalian Genome 10:197-200.
[14] Lamba, P., T. A. Hjalt, and D. J. Bernard. 2008. Novel forms of Paired-like homeodomain transcription factor 2 (PITX2): Generation by alternative translation initiation and mRNA splicing. BMC Molecular Biology 9:31.
[15] Zhang, C., W.-H. Li, A. R. Krainer, and M. Q. Zhang. 2008. RNA landscape of evolution for optimal exon and intron discrimination. Proceedings of the National Academy of Sciences USA 105:5797–5802.
[16] Exodus 31:18, Deuteronomy 9:10
[17] John 8:7,8
[18] Psalm 139:14
Timothy G. Standish, PhD
Senior Scientist, Geoscience Research Institute
A former version of this article was published in German on the magazine Info Vero